Microbial evolution and the spread of infectious disease
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/201812_CBB_Seminar.html
Human Influenza A viruses

Weekly numbers of positive influenza tests in the US by subtype
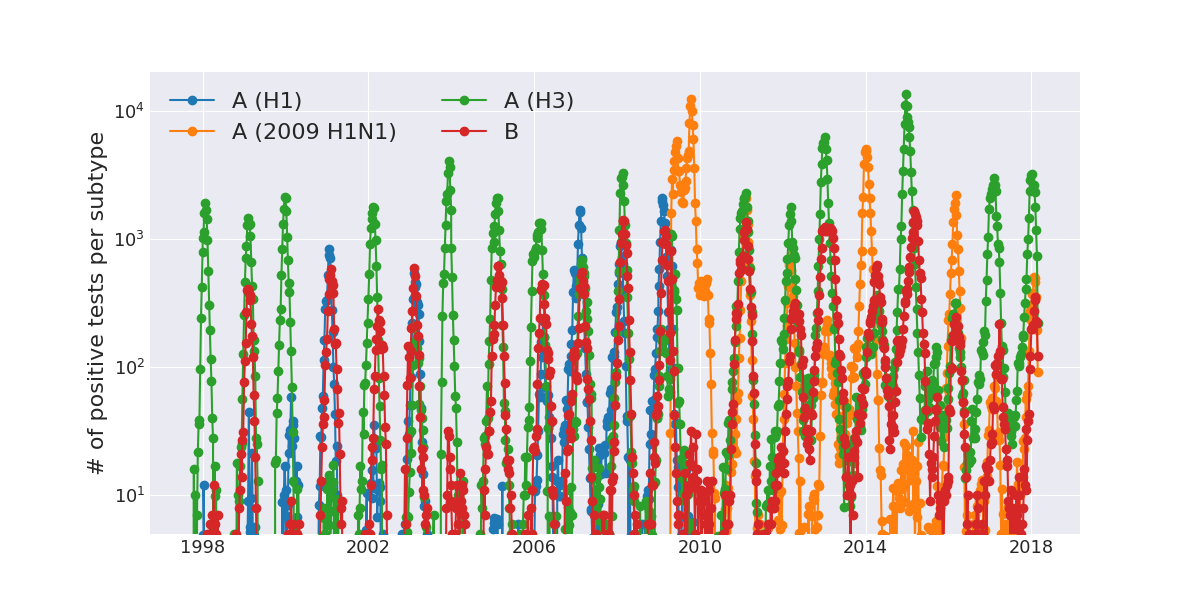 Data by the US CDC
Data by the US CDC



- Influenza virus evolves to avoid human immunity
- Vaccines need frequent updates

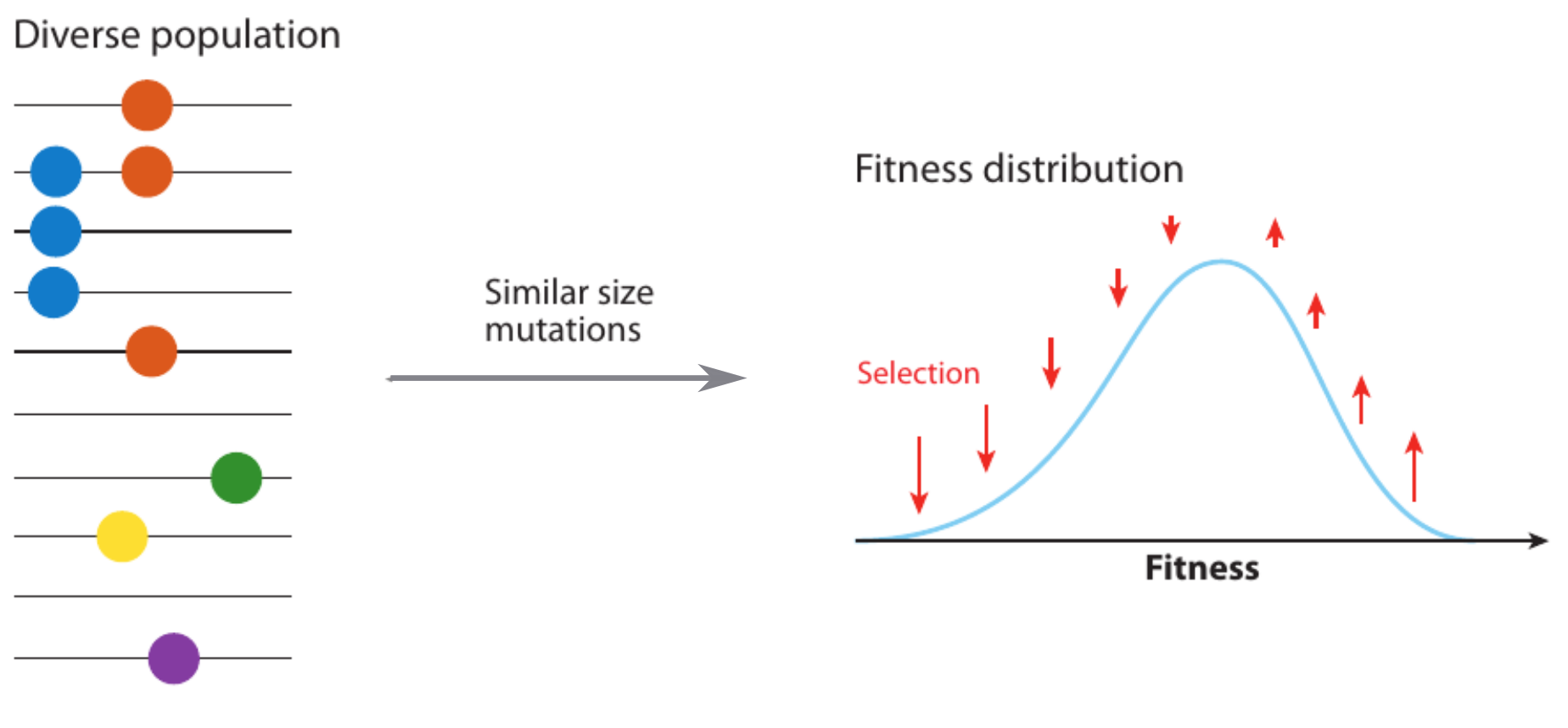
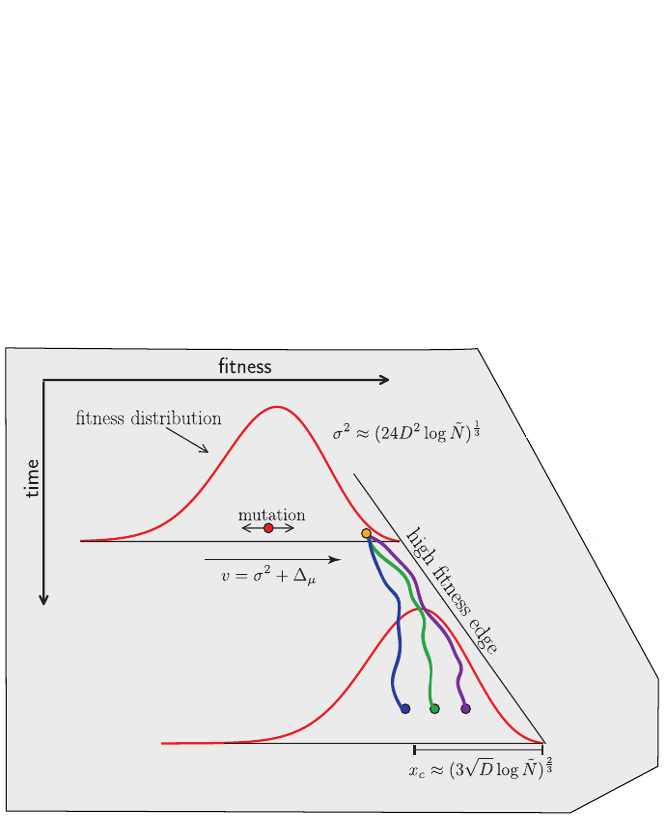
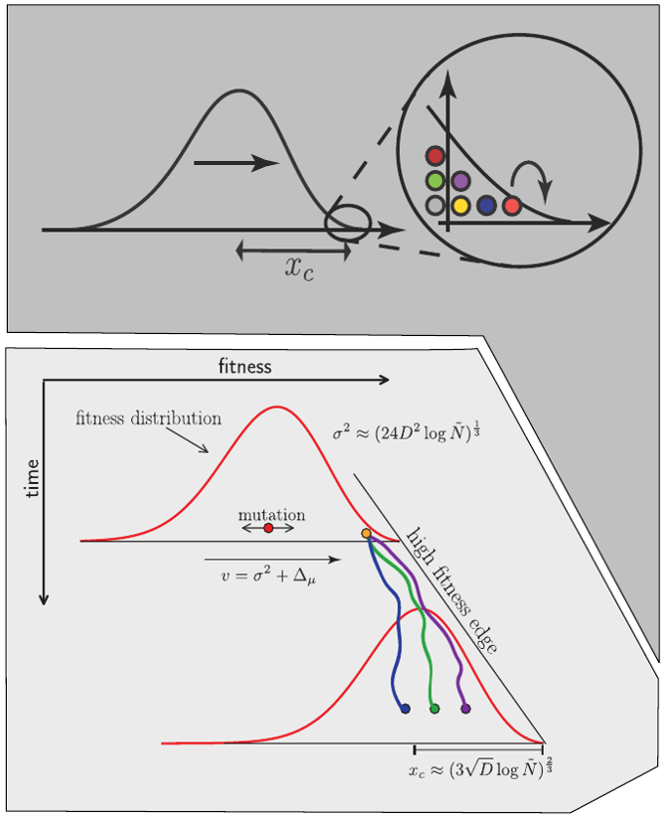
Clonal interference and traveling waves
Typical tree

Bolthausen-Sznitman Coalescent
Bursts in a tree ↔ high fitness genotypes
Can we read fitness of a tree?
Predicting evolution

Given the branching pattern:
- can we predict fitness?
- pick the closest relative of the future?
Fitness inference from trees
$$P(\mathbf{x}|T) = \frac{1}{Z(T)} p_0(x_0) \prod_{i=0}^{n_{int}} g(x_{i_1}, t_{i_1}| x_i, t_i)g(x_{i_2}, t_{i_2}| x_i, t_i)$$
RN, Russell, Shraiman, eLife, 2014
Prediction of the dominating H3N2 influenza strain
- no influenza specific input
- how can the model be improved? (see model by Luksza & Laessig)
- what other context might this apply?
nextstrain.org
joint work with Trevor Bedford & his lab
HIV-1 evolution within one individual
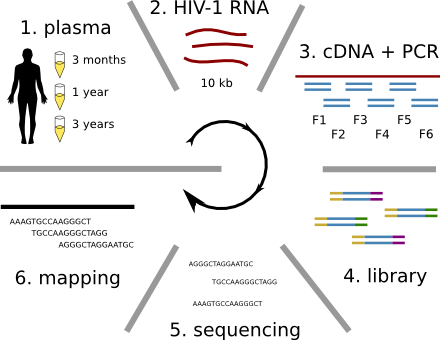

Population sequencing to track all mutations above 1%
Bacterial evolution
- every genome contains a different set of genes
- horizontal transfer
- genome rearrangements
- variable divergence along the genome
- important parts sit on plasmids,
flanked by repetitive sequence - short read assemblies tend to be fragmented
- annotations of variable quality

panX by Wei Ding
- pan-genome identification pipeline
- phylogenetic analysis of each orthologous cluster
- detect associations with phenotypes
- fast: analyze hundreds of genomes in a few hours
- github.com/neherlab/pan-genome-analysis
HIV acknowledgments







- Fabio Zanini
- Jan Albert
- Johanna Brodin
- Christa Lanz
- Göran Bratt
- Lina Thebo
- Vadim Puller



Influenza and Theory acknowledgments




- Boris Shraiman
- Colin Russell
- Trevor Bedford
- Oskar Hallatschek



Acknowledgments






- Trevor Bedford
- Colin Megill
- Pavel Sagulenko
- Sidney Bell
- James Hadfield
- Wei Ding
- Emma Hodcroft
- Sanda Dejanic



