Evolution and spread of SARS-CoV-2
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/202006_KITP.html
Acknowledgements
Trevor Bedford and his lab -- terrific collaboration since 2014

James Hadfield, Emma Hodcroft and Tom Sibley have led the recent development
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID


 Data summarized by Ian MacKay
Data summarized by Ian MacKay
 Data summarized by Ian MacKay
Data summarized by Ian MacKay
 Data summarized by Ian MacKay
Data summarized by Ian MacKay
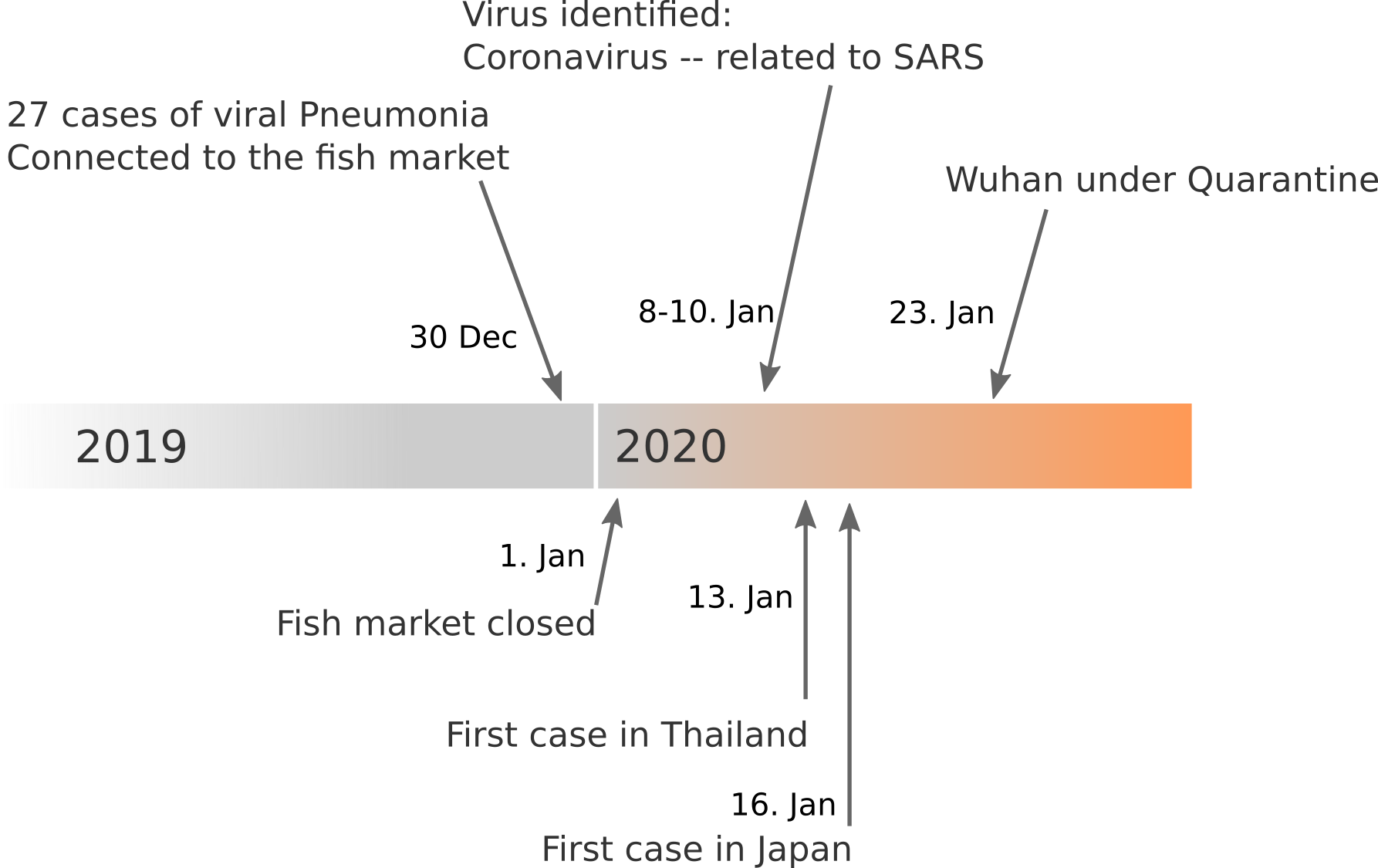 Data summarized by Ian MacKay
Data summarized by Ian MacKay
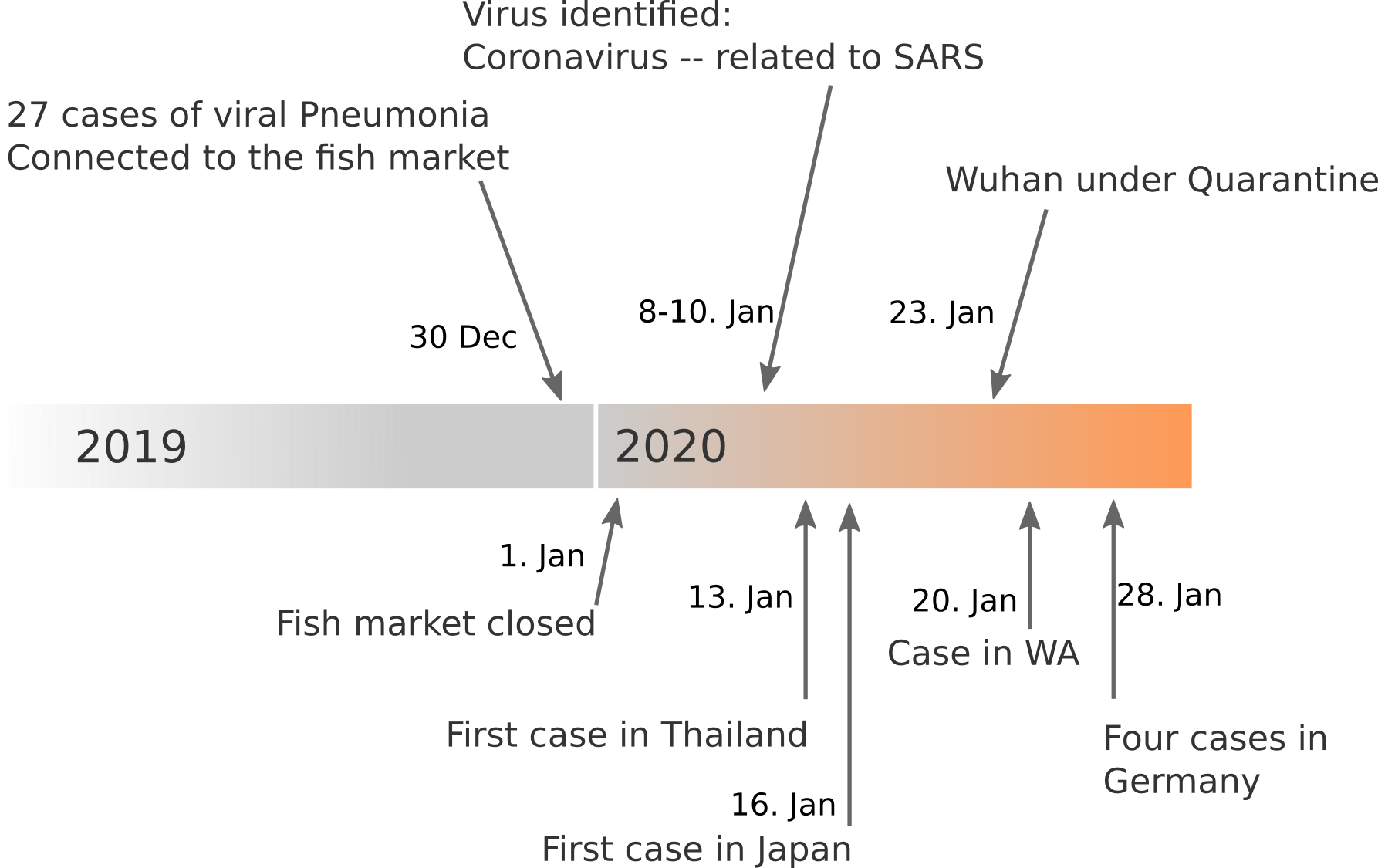 Data summarized by Ian MacKay
Data summarized by Ian MacKay
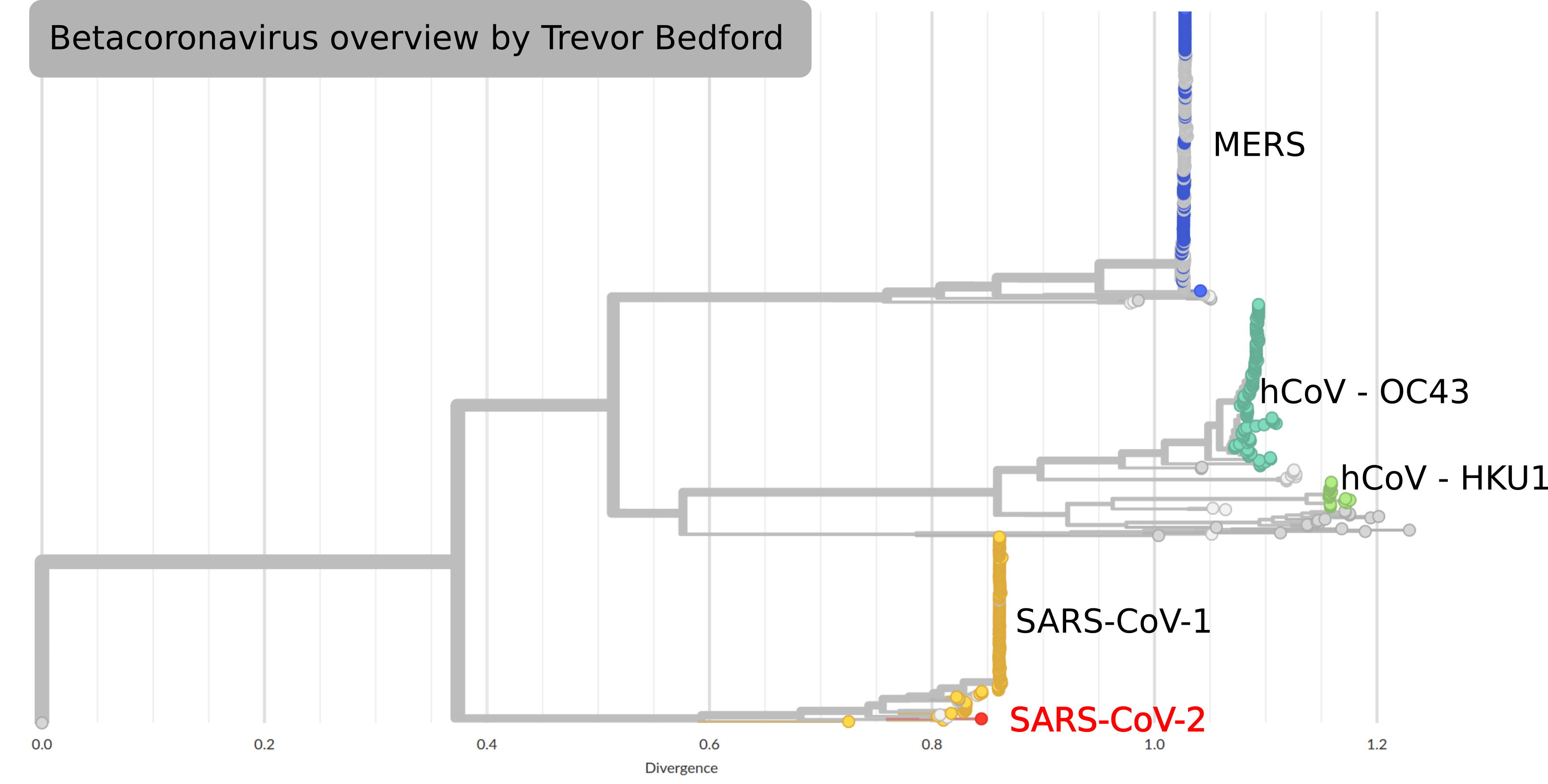
SARS-CoV-2 and its relatives
- SARS-CoV-2 is in the same family as SARS-CoV-1, MERS and two seasonal coronavirus
- The latter cause mild disease, the former severe
- SARS-CoV-2 spreads more easily than SARS-CoV-1 or MERS
- Relatives are found in many different animals
- Closest known relative is a virus isolated from bats
(RaTG13, approx 96% identical)
Tracking diversity and spread of SARS-CoV-2 in nextstrain
Virus genomes change rapidly through time
A/Brisbane/100/2014
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
A/Brisbane/1000/2015
GGATAATTCTATTAACCATGAAGACTATTATTGCTTT...
A/Brisbane/1/2017
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
... tens of thousands of sequences...
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
A/Brisbane/1000/2015
GGATAATTCTATTAACCATGAAGACTATTATTGCTTT...
A/Brisbane/1/2017
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
... tens of thousands of sequences...
Genome sequences record the spread of pathogens




in SARS-CoV-2 a new mutation accumulates about every 2 weeks
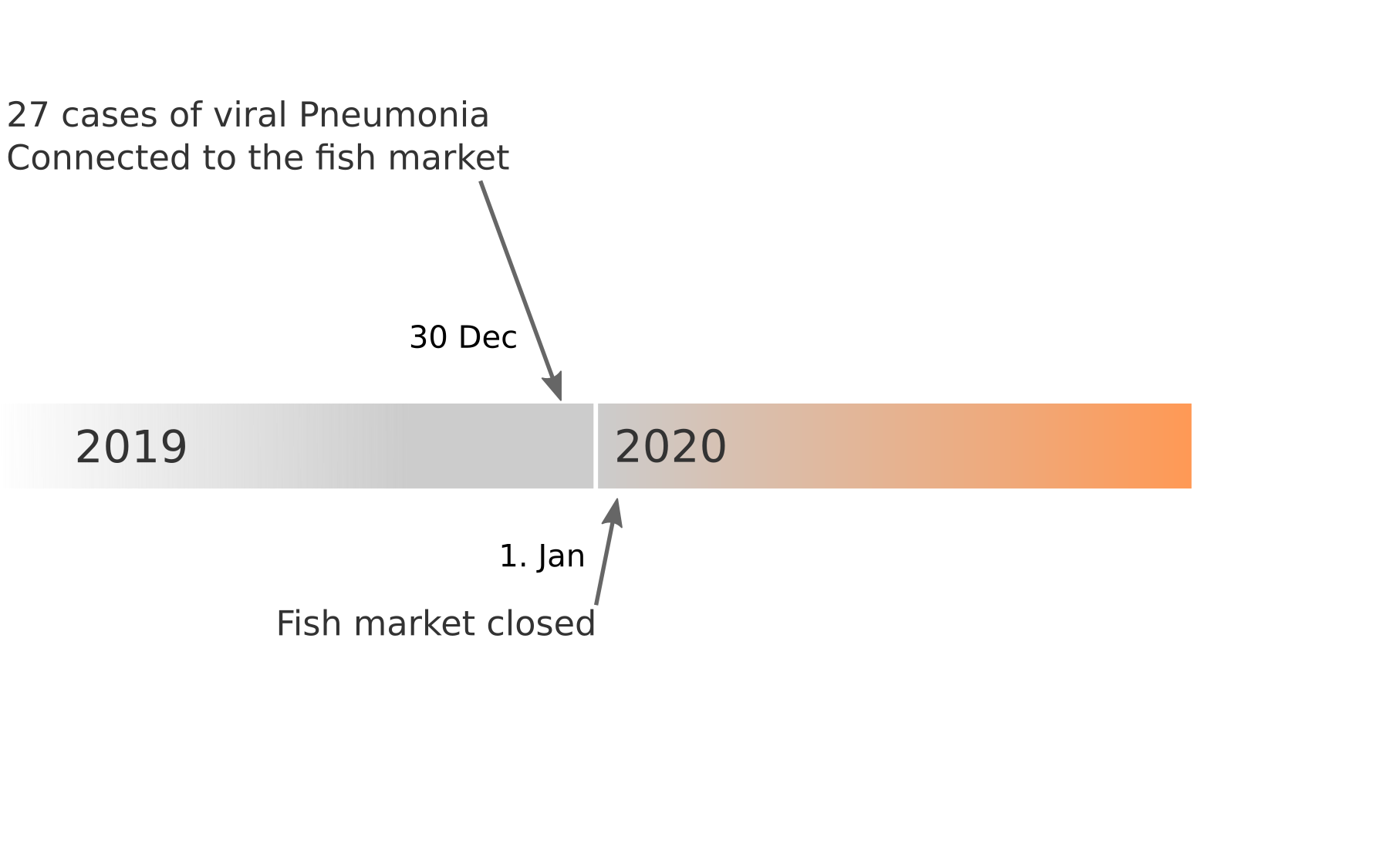
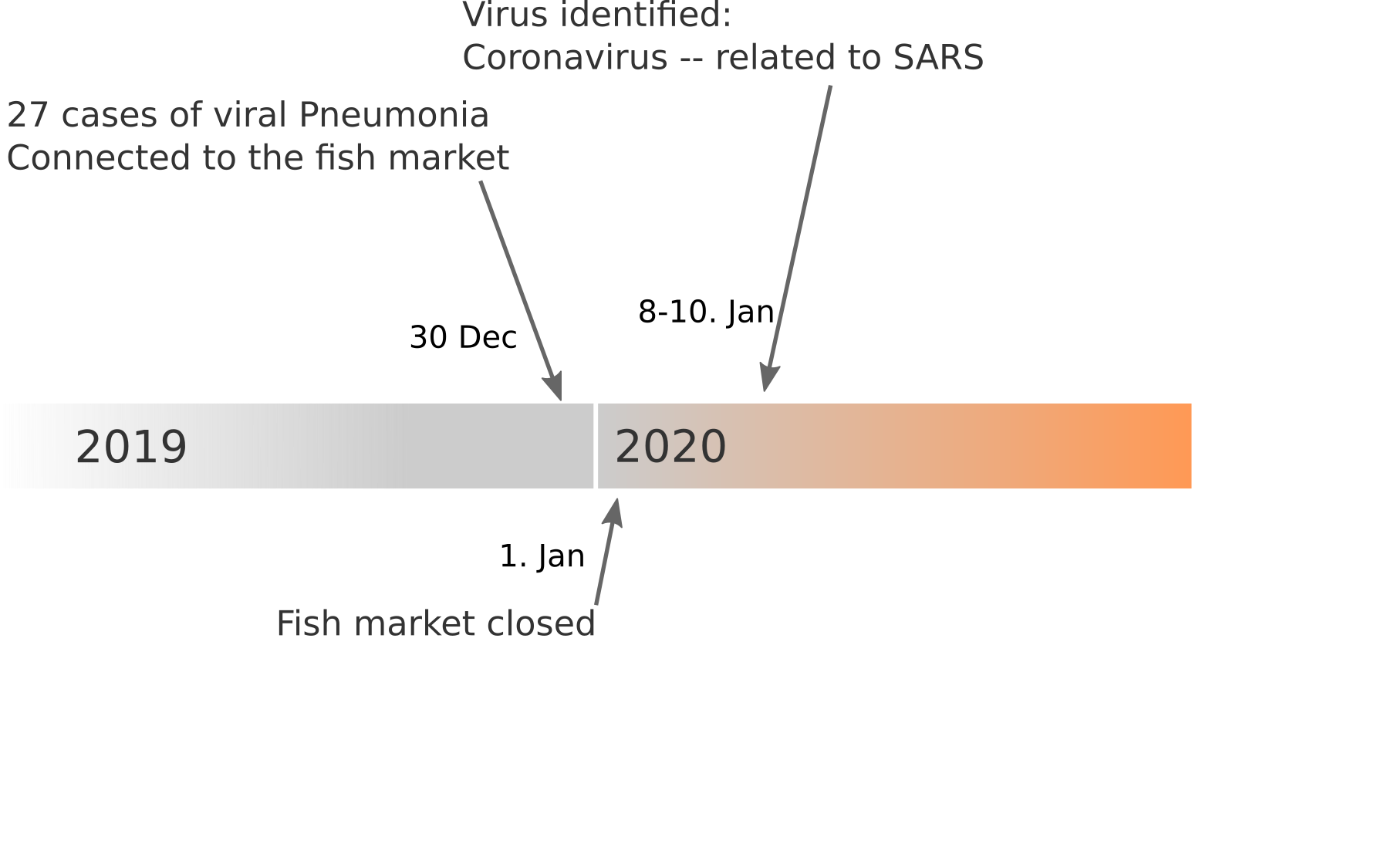
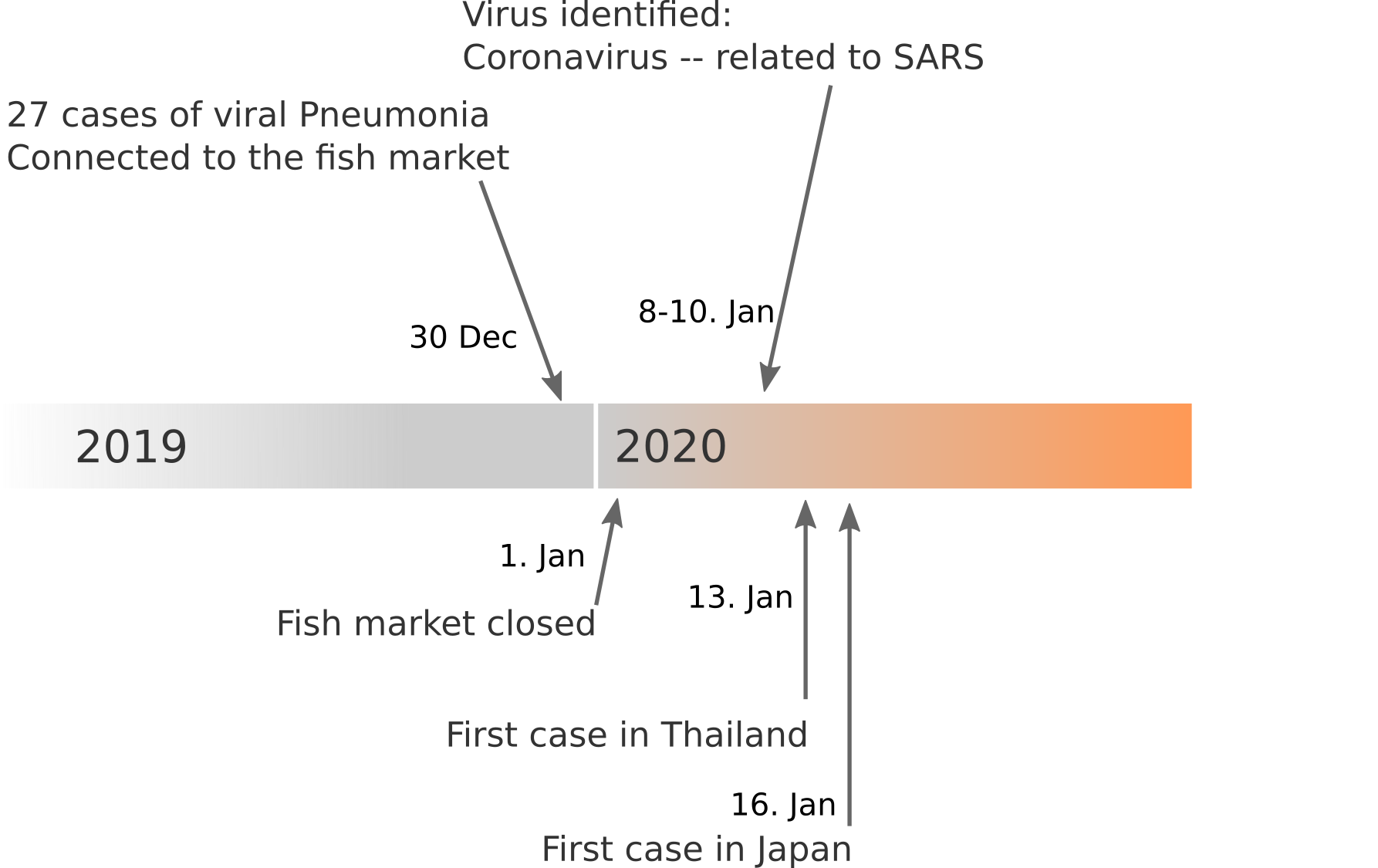
Available data on Jan 26
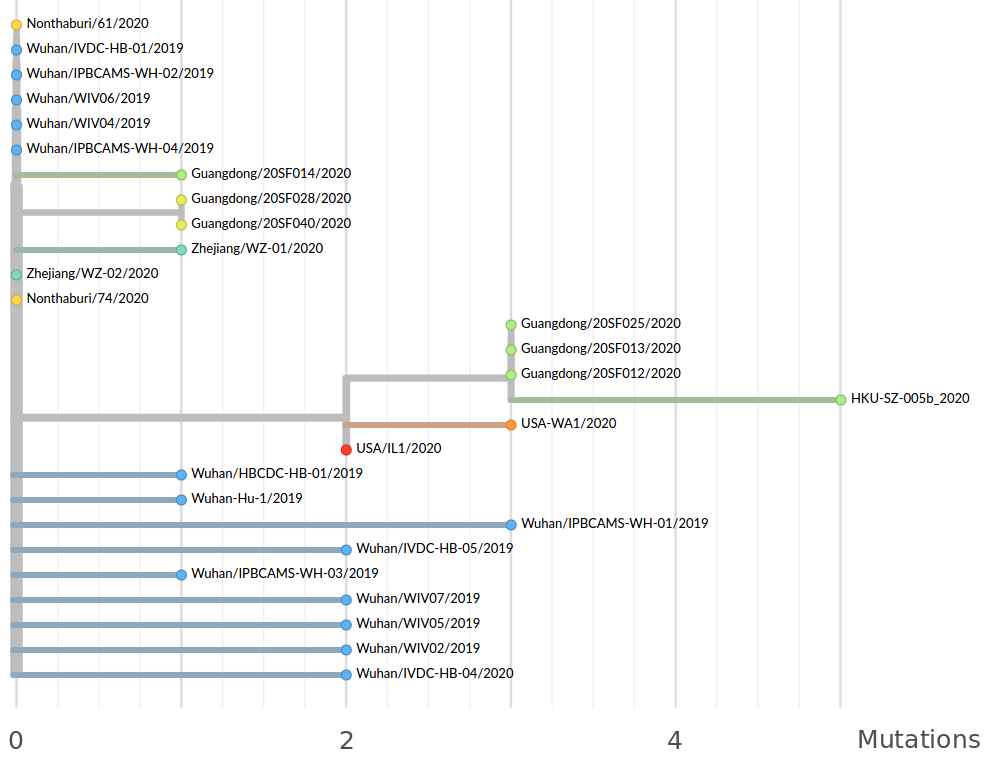
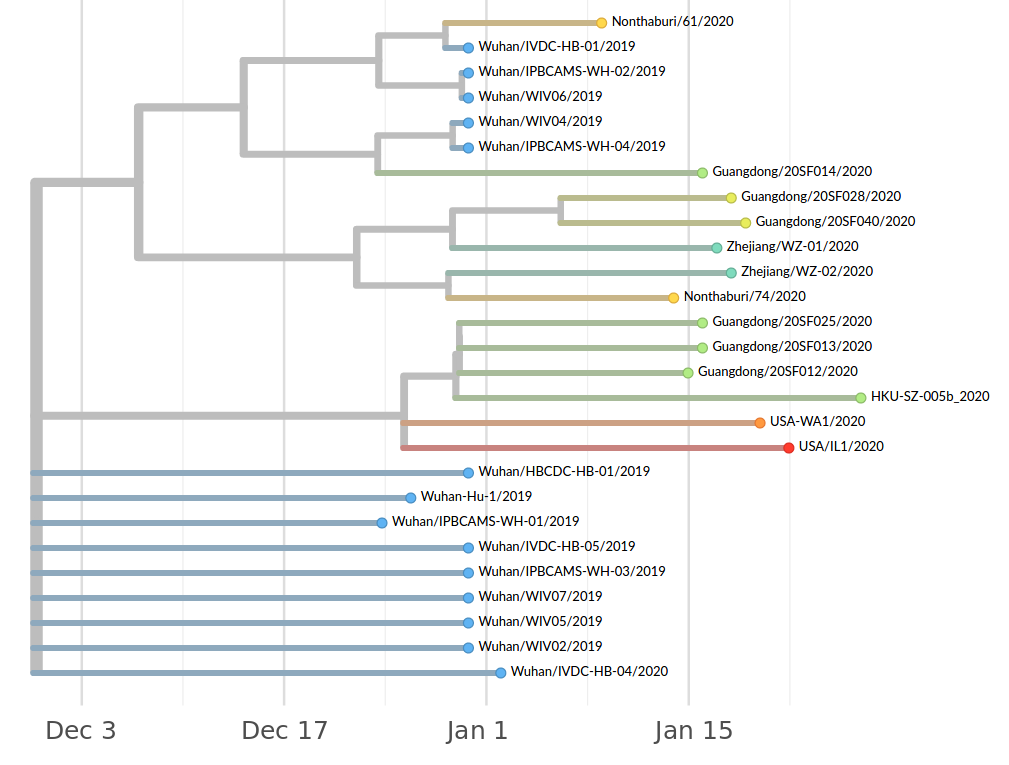
Early genomes differed by only a few mutations, suggesting very recent emergence
Early lessons from SARS-CoV-2 genomes (mid Jan)
- The outbreak originated from one source
→ not repeated zoonoses from a diverse reservoir. - The common ancestor of all samples was Nov - early Dec 2019.
- Family clusters showed up as identical genomes (expected)
- A second clade emerged that will continue to spread
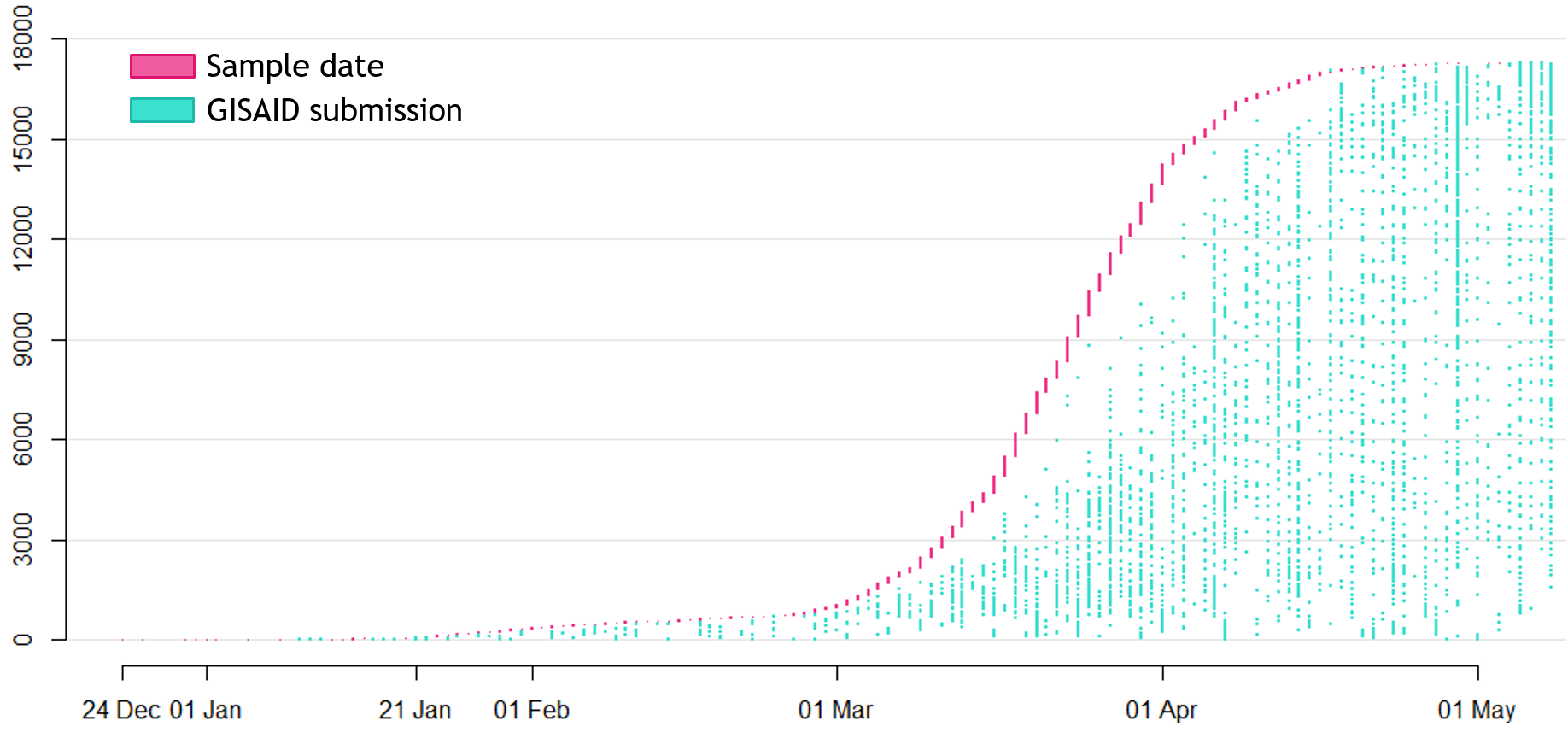
→ the closest to "real-time" we have experienced so far...
Figure by James Hadfield/Emma HodcroftAvailable data on June 4
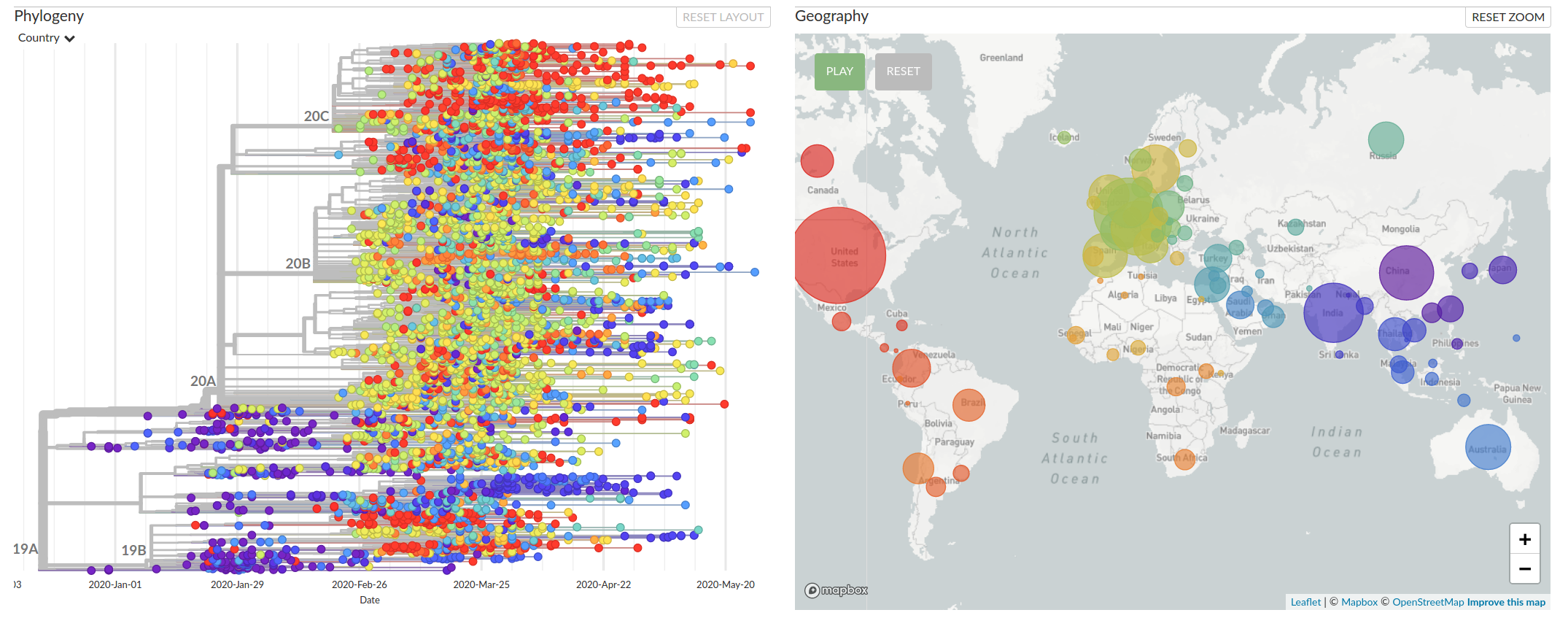
The virus has reached most places where humans live
Available data on June 4
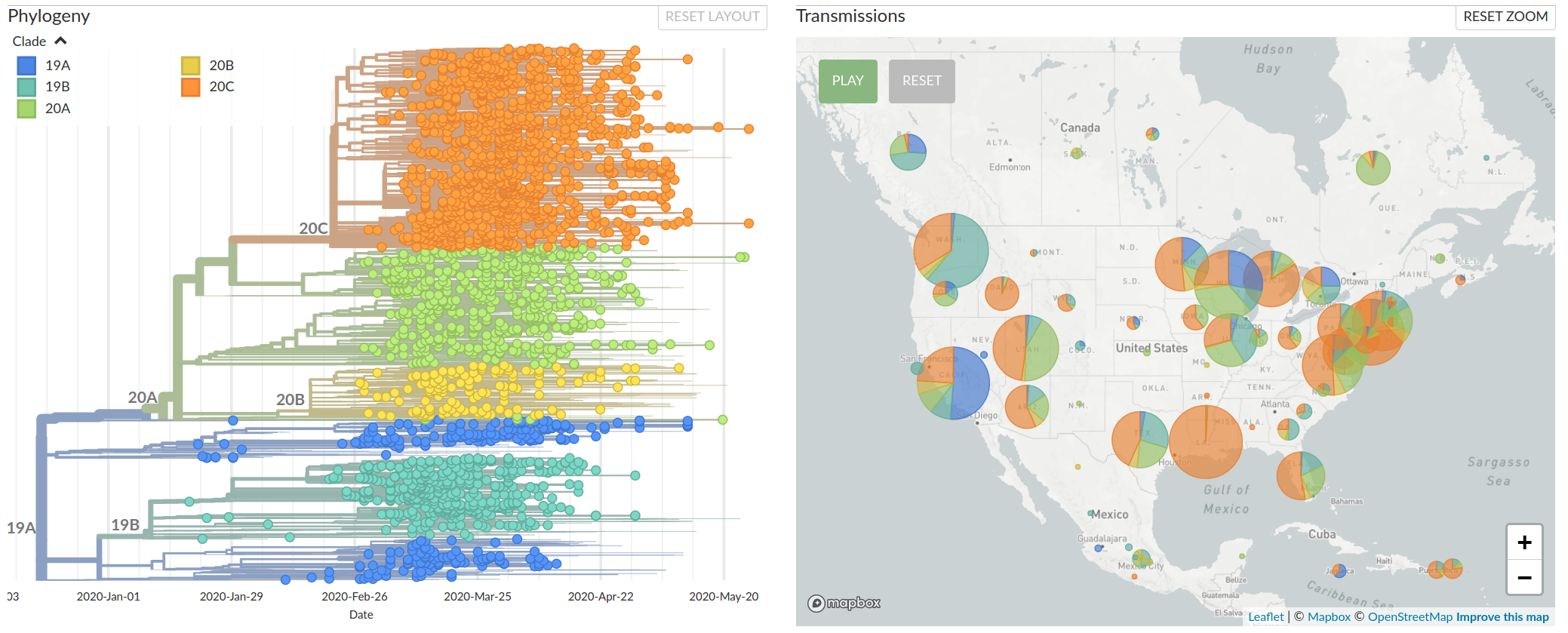
Different clades (virus variants) dominate in different parts of the US
Available data on June 4
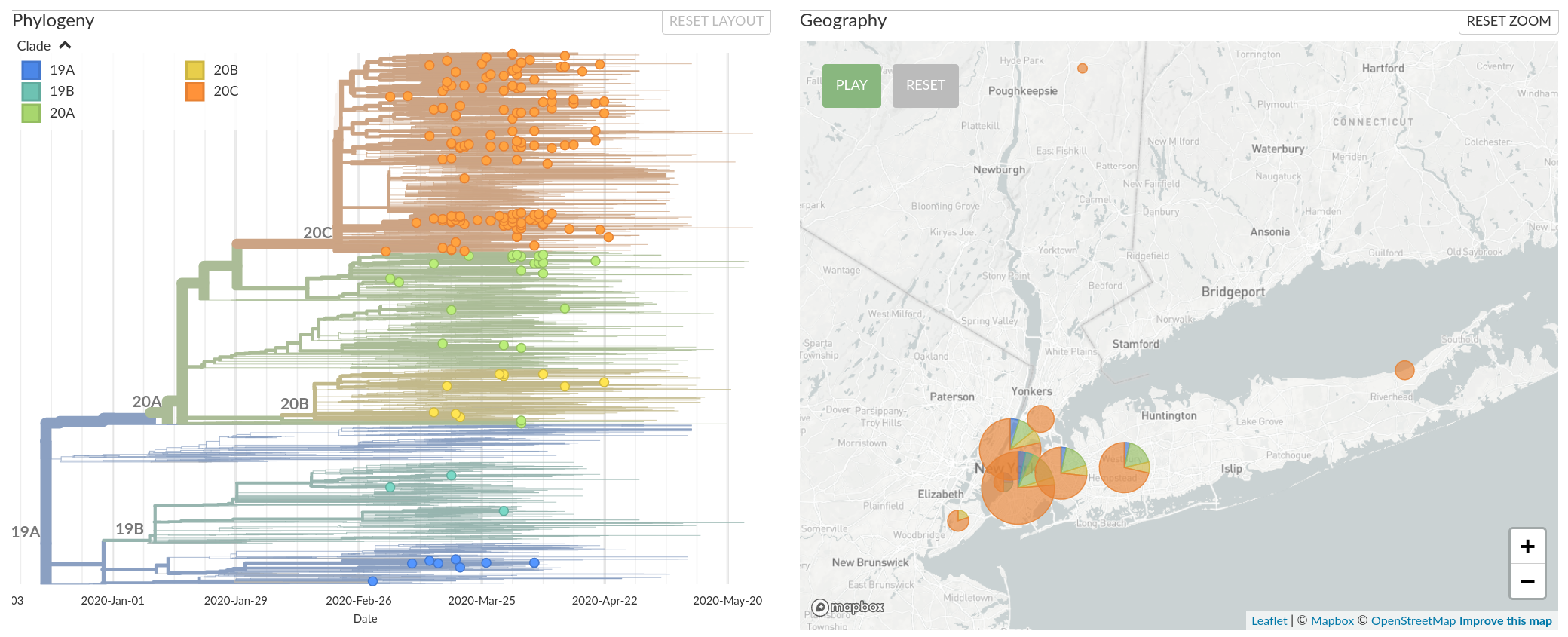
Data from New York suggests viral diversity
Available data on June 4
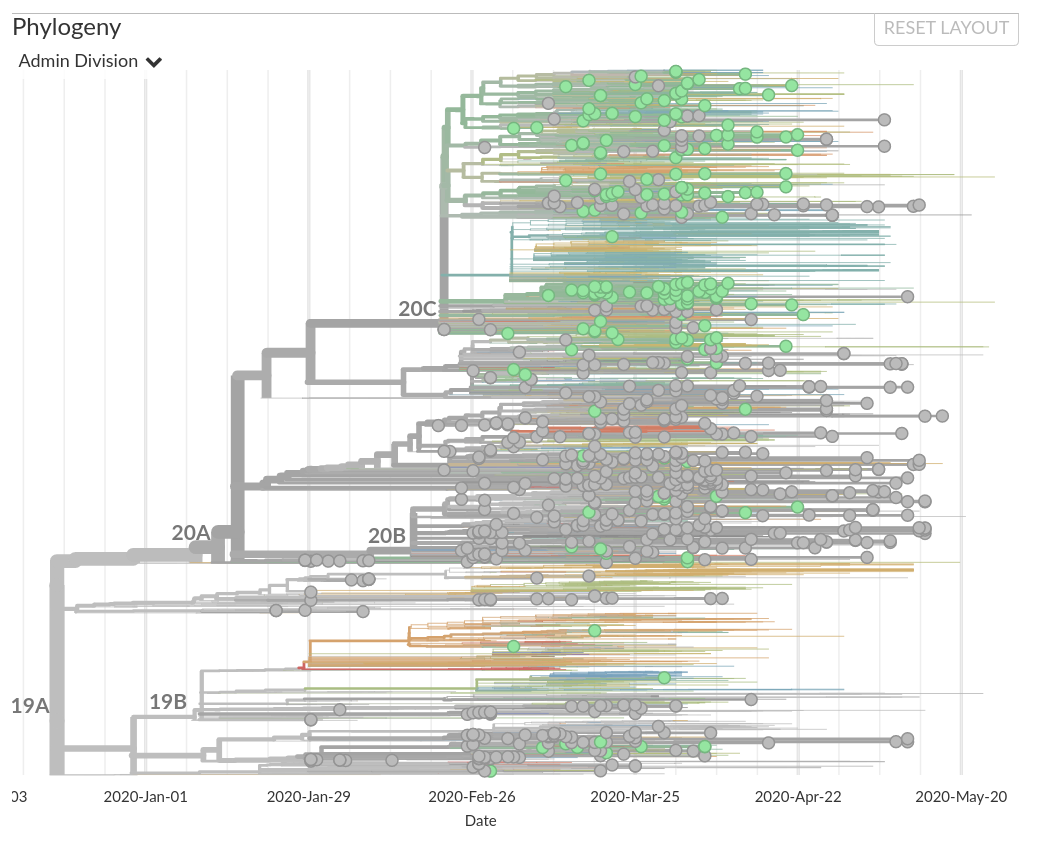
The outbreak in New York was likely seeded by introductions from Europe
Genomes reveal how the virus moves around the globe
Tracing the origins of samples from Iceland
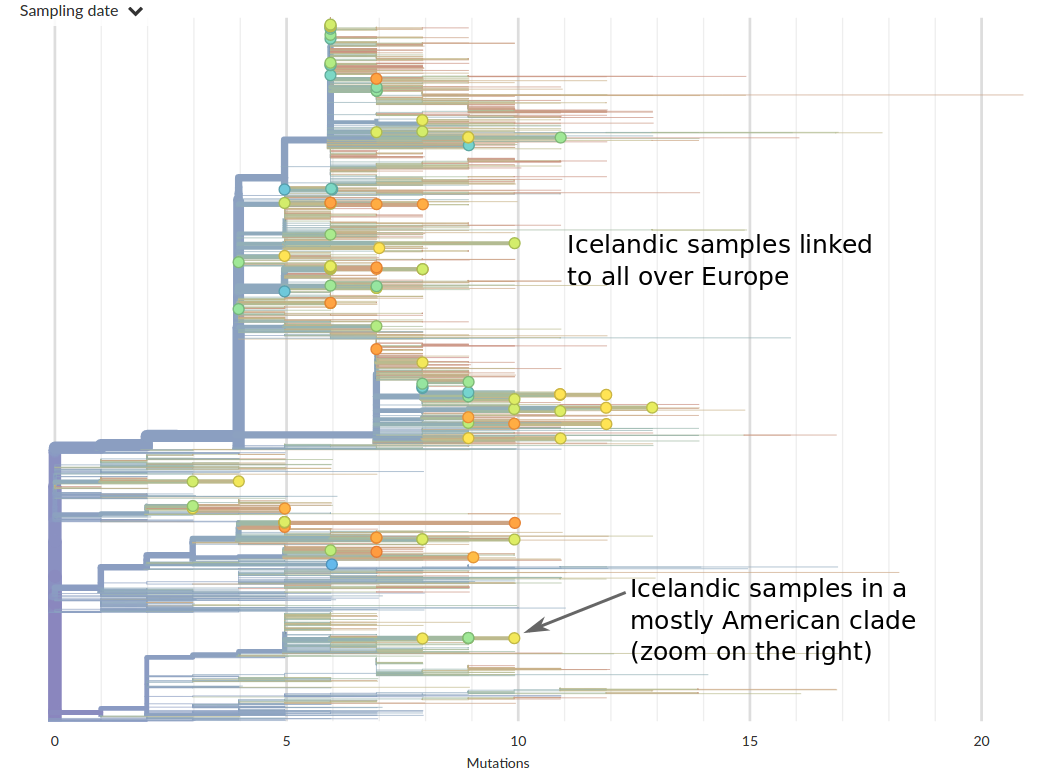
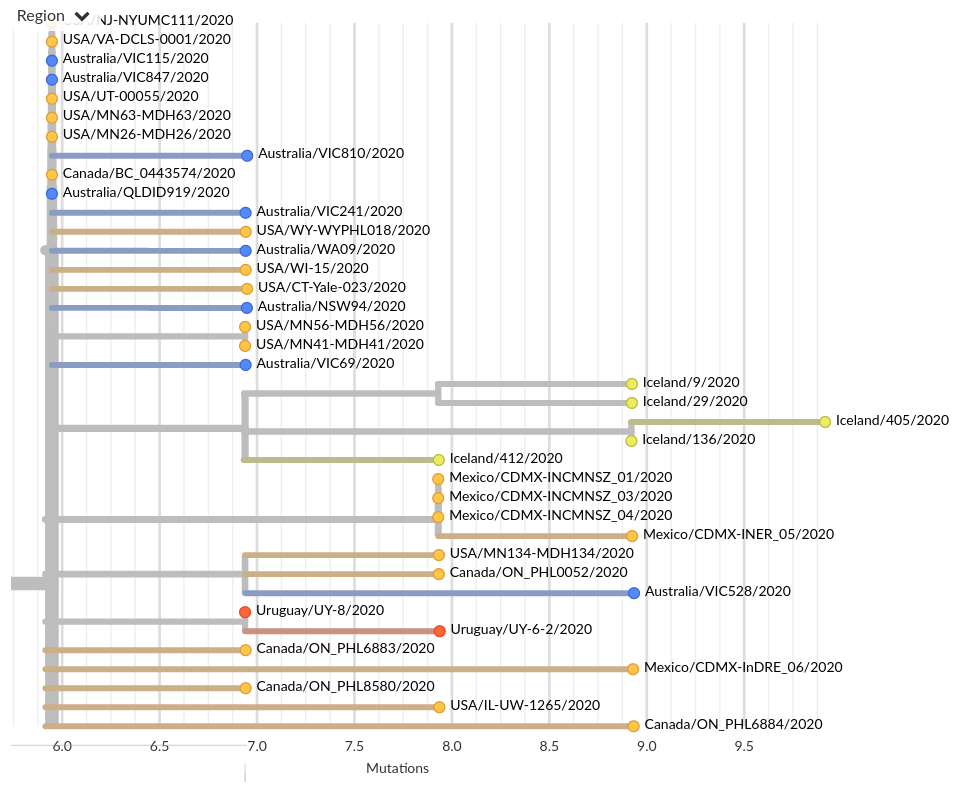
SARS-CoV-2 is remarkably mixed geographically -- genomes connect outbreaks in different places
Current take-home from SARS-CoV-2 genomes
- SARS-CoV-2 was disseminated widely before travel restrictions
→ Border closures had little effect (maybe if earlier) - The virus typically spread locally before it was noticed
(while testing focussed on people with travel history to hotspots) - Temporal resolution by mutation is about 4 weeks
→ too low to resolve direct transmissions or directionality
→ sufficient to connect outbreaks and identify clusters - Currently no solid evidence for mutations with functional significance
see Sergei Pond's analysis of signals of positive selection
The history of nextstrain -- KITP in summer 2014
Nextstrain started at the KITP/superbugs14 program
- Boris Shraiman and I had worked on prediction of influenza virus
- Trevor Bedford had the vision of an "always-on server [..] predicting clade trajectories"
- In early 2015 we had a prototype for influenz virus (nextflu)
- Rapidly diversified to cover Ebola Virus and MERS CoV
- Rebranded as nextstrain


Trevor Bedford, FHCRC Seattle
Fighting misinformation
Conspiracy theories and sensationalist research
WRONG:
- COVID-19 has been with us for much longer
Genetic diversity suggests recent emergence, respiratory samples from last year are PCR negative - Origin are snakes (snake-flu)
debunked by Kristian Andersen - Recombinant with HIV
debunked by Trevor Bedford
LIKELY WRONG:
- Prevalence is high, the mortality is low
False positive rates are not accounted for correctly, unrepresentative study populations - Some strains are more severe than others
confounders not accounted for - Some strains are adapted to different parts of the world
confounders not accounted for - many more....
Colorful trees are easily misinterpreted...
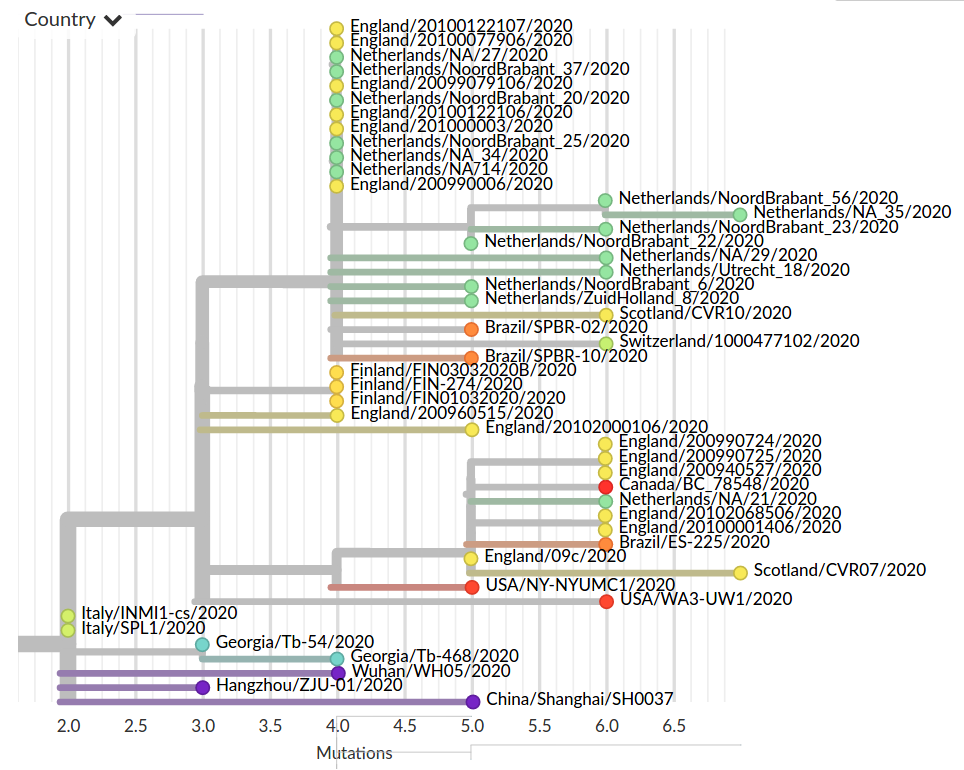
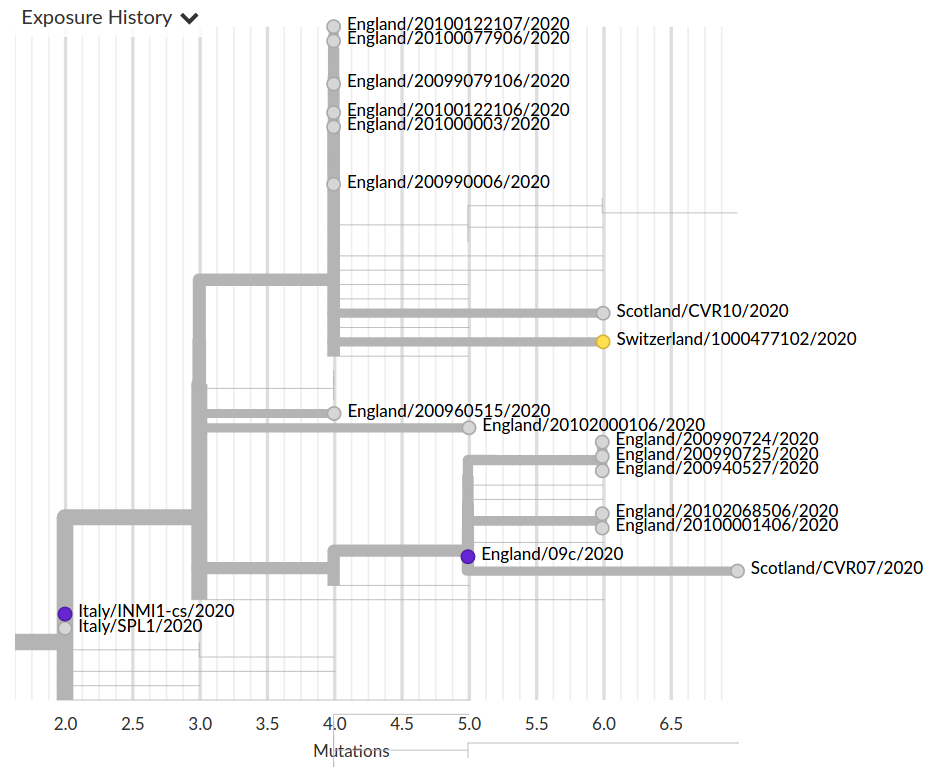
Low genetic diversity combined with very biased sampling → no directionality can be inferred
Acknowledgements
Trevor Bedford and his lab -- terrific collaborations since 2014

James Hadfield, Emma Hodcroft, and Tom Sibley have led the recent development
People at the frontlines: health care staff and everybody else who keeps things going...
Data we analyze are contributed by scientists all over the world
Data are shared and curated by GISAID


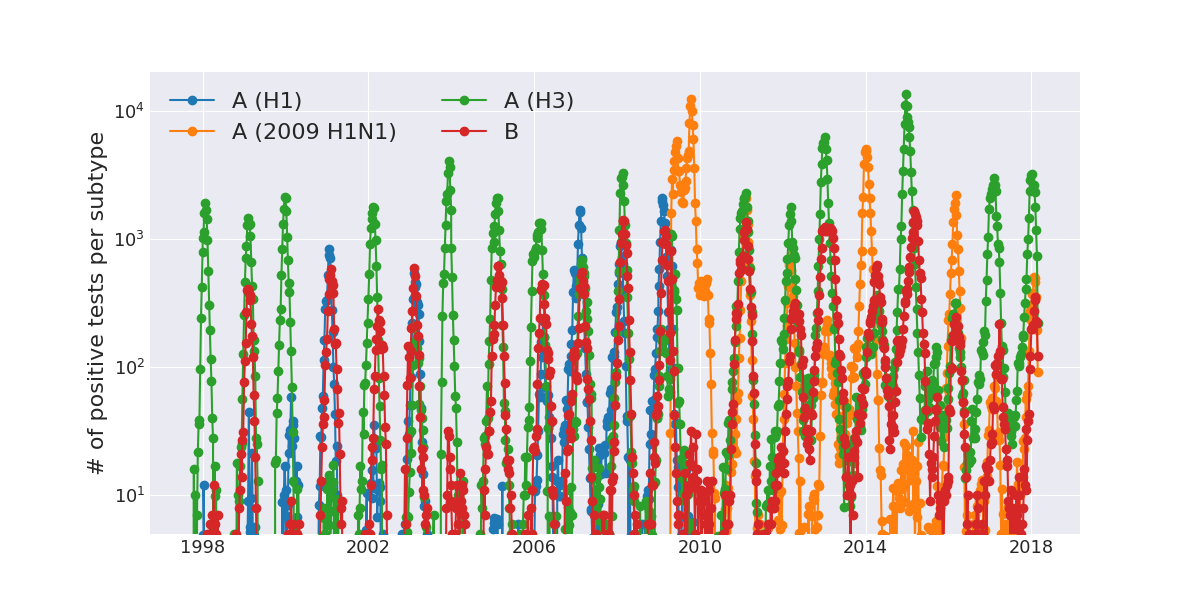
Seasonal variation in infectivity
Seasonal incidence of influenza viruses
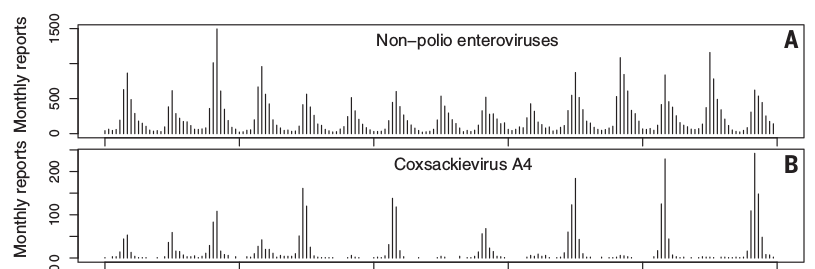
Seasonal incidence of enteroviruses
Potential seasonal drivers
- humidity (e.g. dry air increases time before droplets fall down)
- temperature
- UV light
- behavior (e.g. time spent indoors, ventilation, heating)
Exact nature of forcing not relevant for this talk...
2009 pandemic influenza -- UK
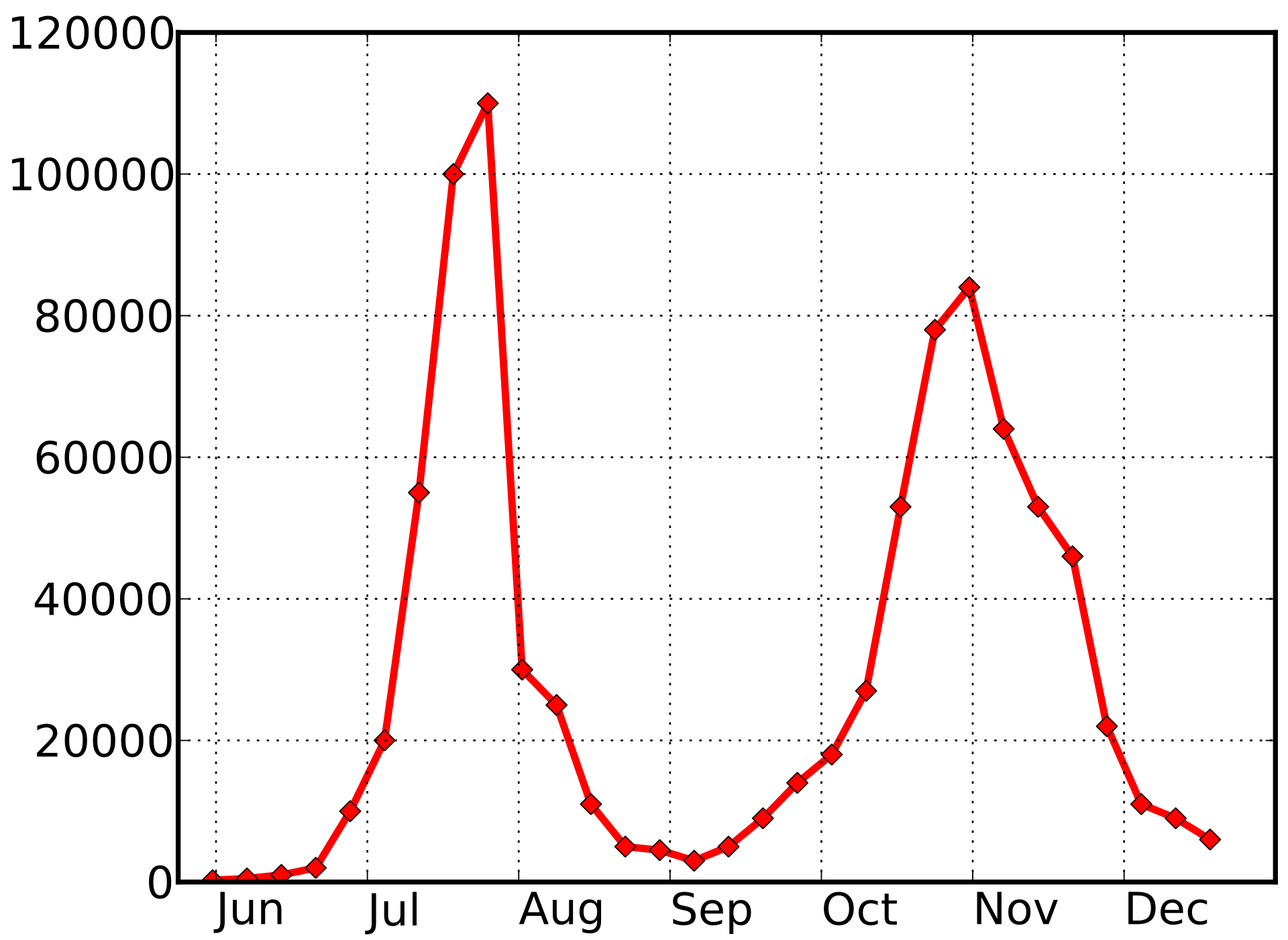
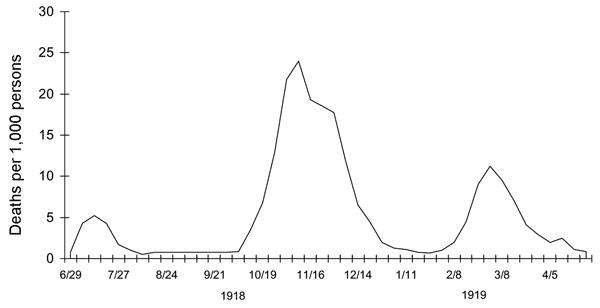
1918 influenza --- UK
 by Trevor Bedford
by Trevor Bedford
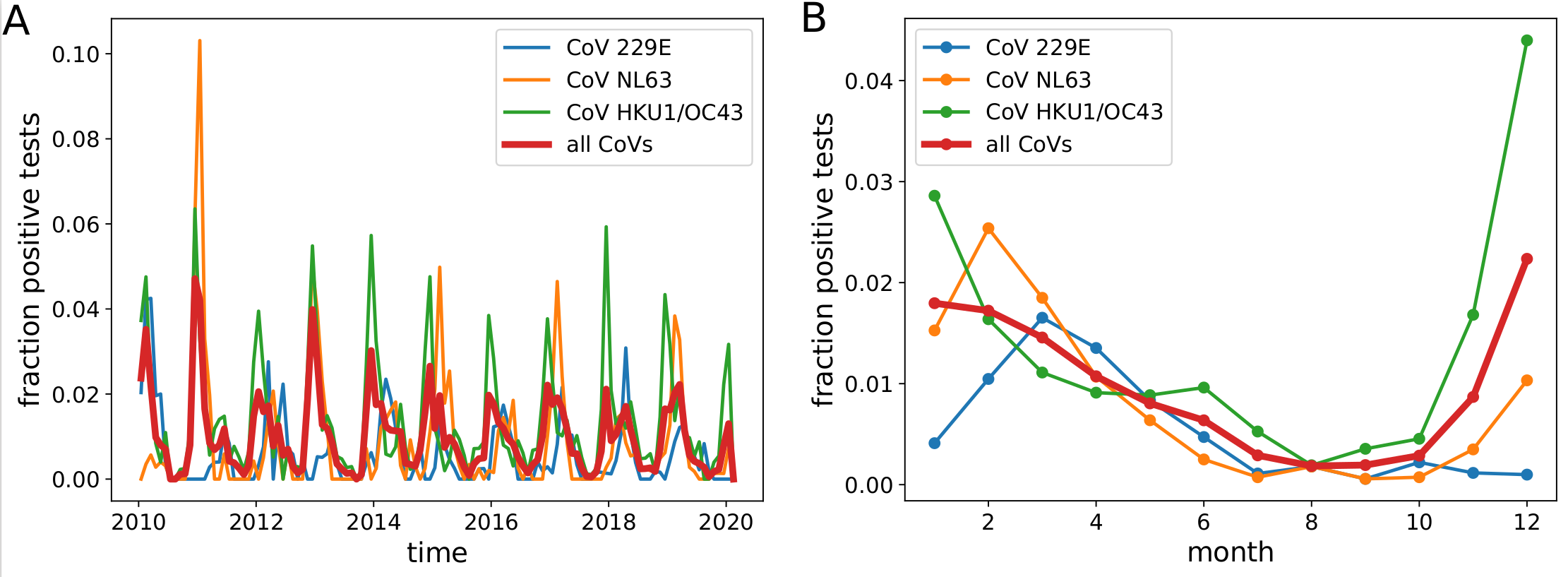
Human corona viruses have pronounced seasonal prevalence (Sweden)
SIR model with seasonal forcing
People import new infections at rate $\nu$
Strong seasonality can be generated through strong forcing or resonance
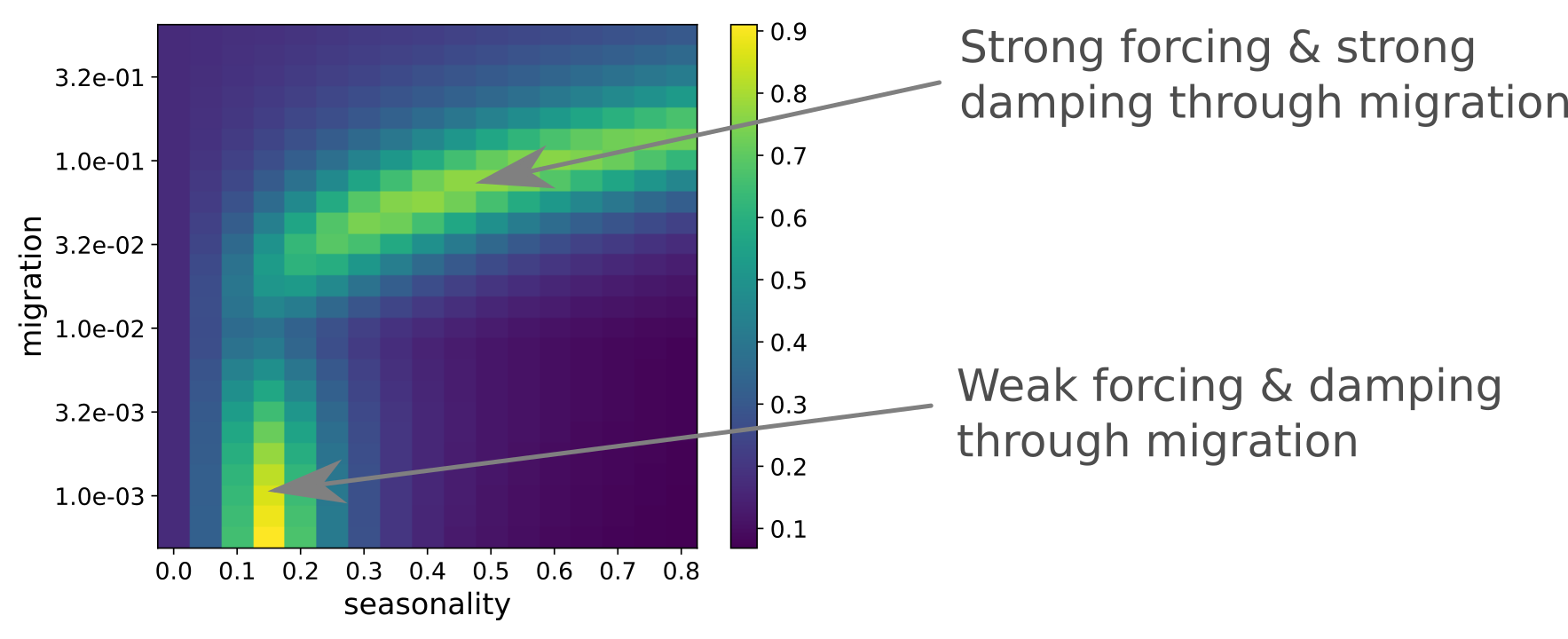
Yellow: good fit -- Blue: poor fit
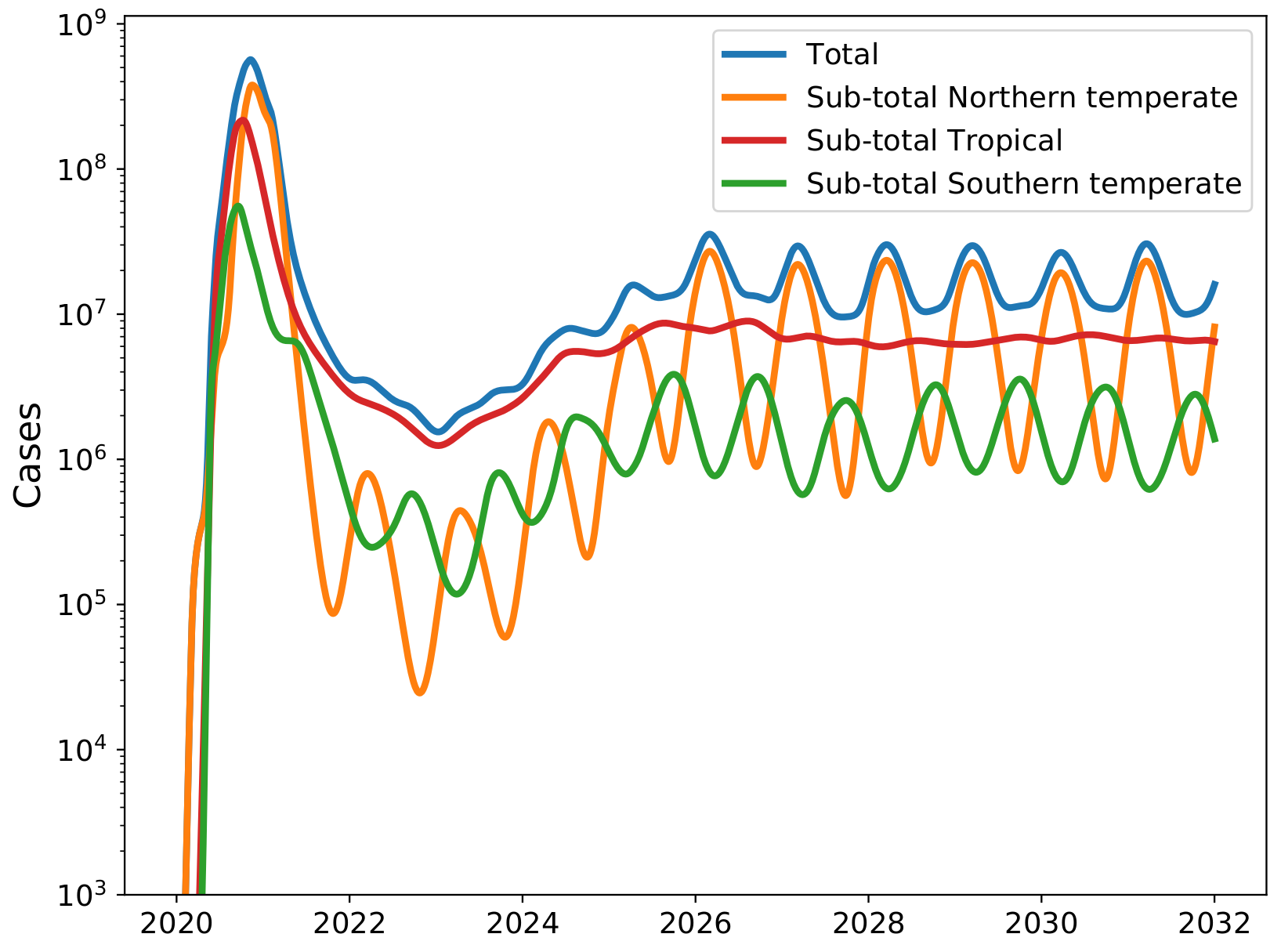
Seasonal forcing and the SARS-CoV-2 pandemic
Counter-factual scenario without strong social distancing!
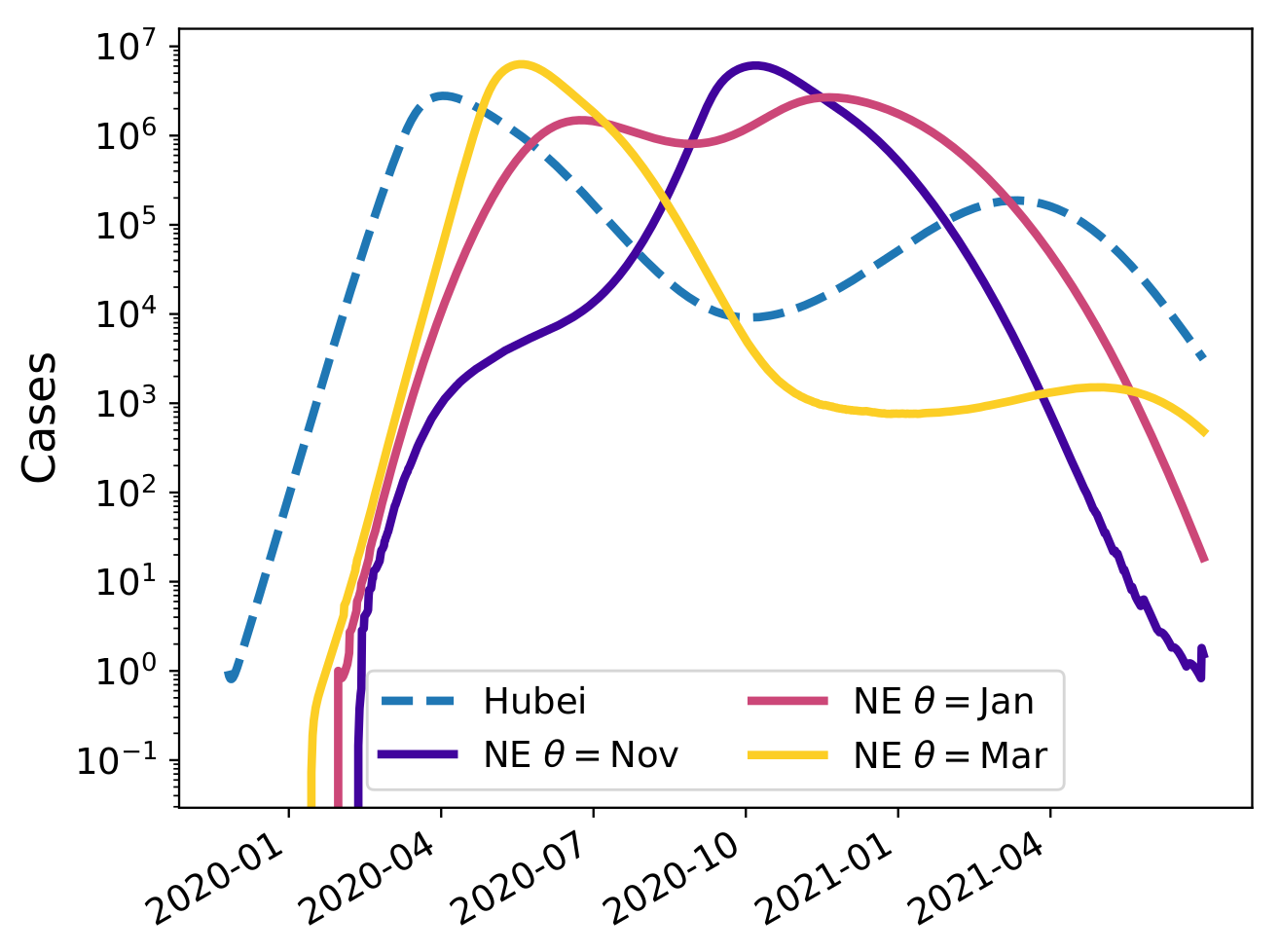
Seasonal forcing and the SARS-CoV-2 pandemic
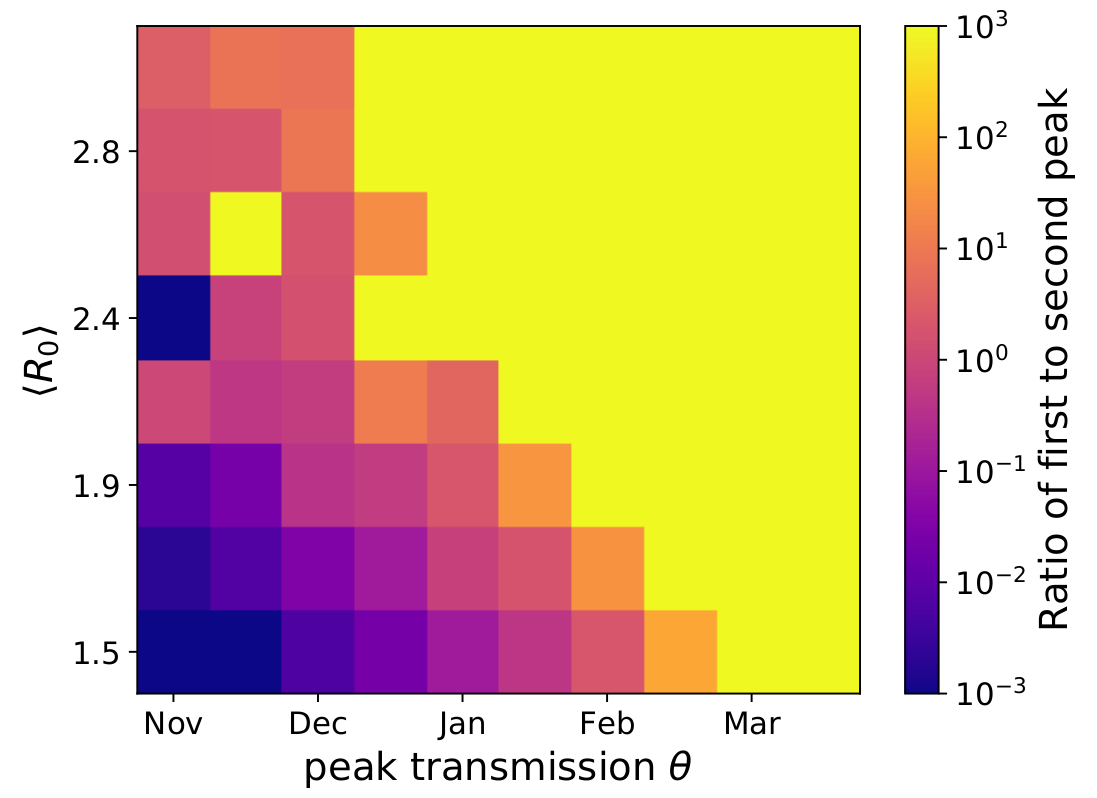
Potential transition to an endemic seasonal virus