Tracking and predicting the evolution of human RNA viruses
Richard Neher
Biozentrum & SIB, University of Basel
slides at neherlab.org/202010_rockefeller.html
Human seasonal influenza viruses

Positive tests for influenza in the USA by week
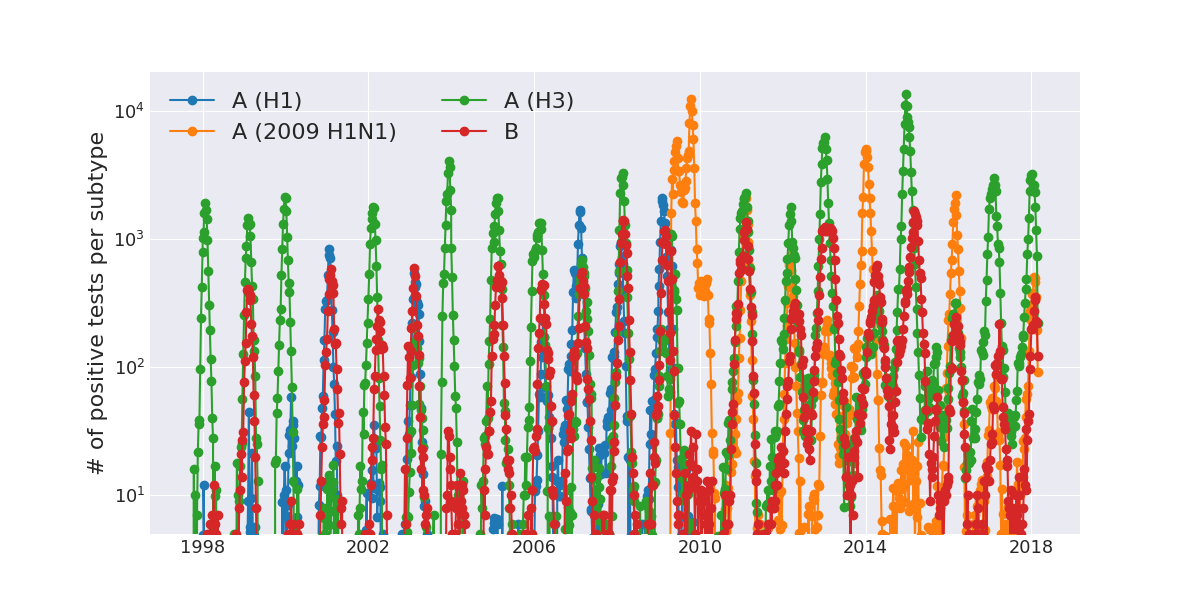 Data by the US CDC
Data by the US CDC
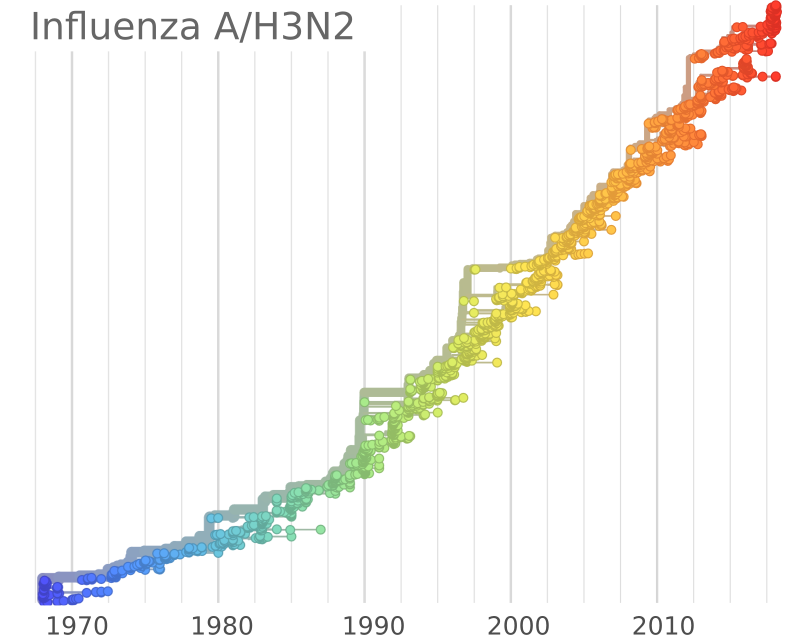
Sequences record the spread of pathogens




Mutations accumulate at a rate of $10^{-5}$ per site and day!



- Influenza viruses evolve to avoid human immunity
- Vaccines need frequent updates

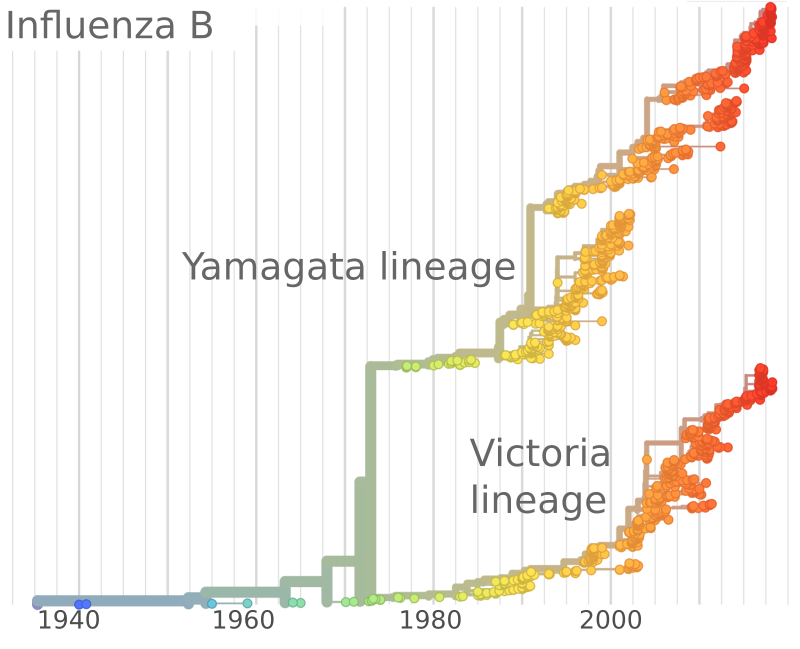
Influenza B viruses have split into two lineages
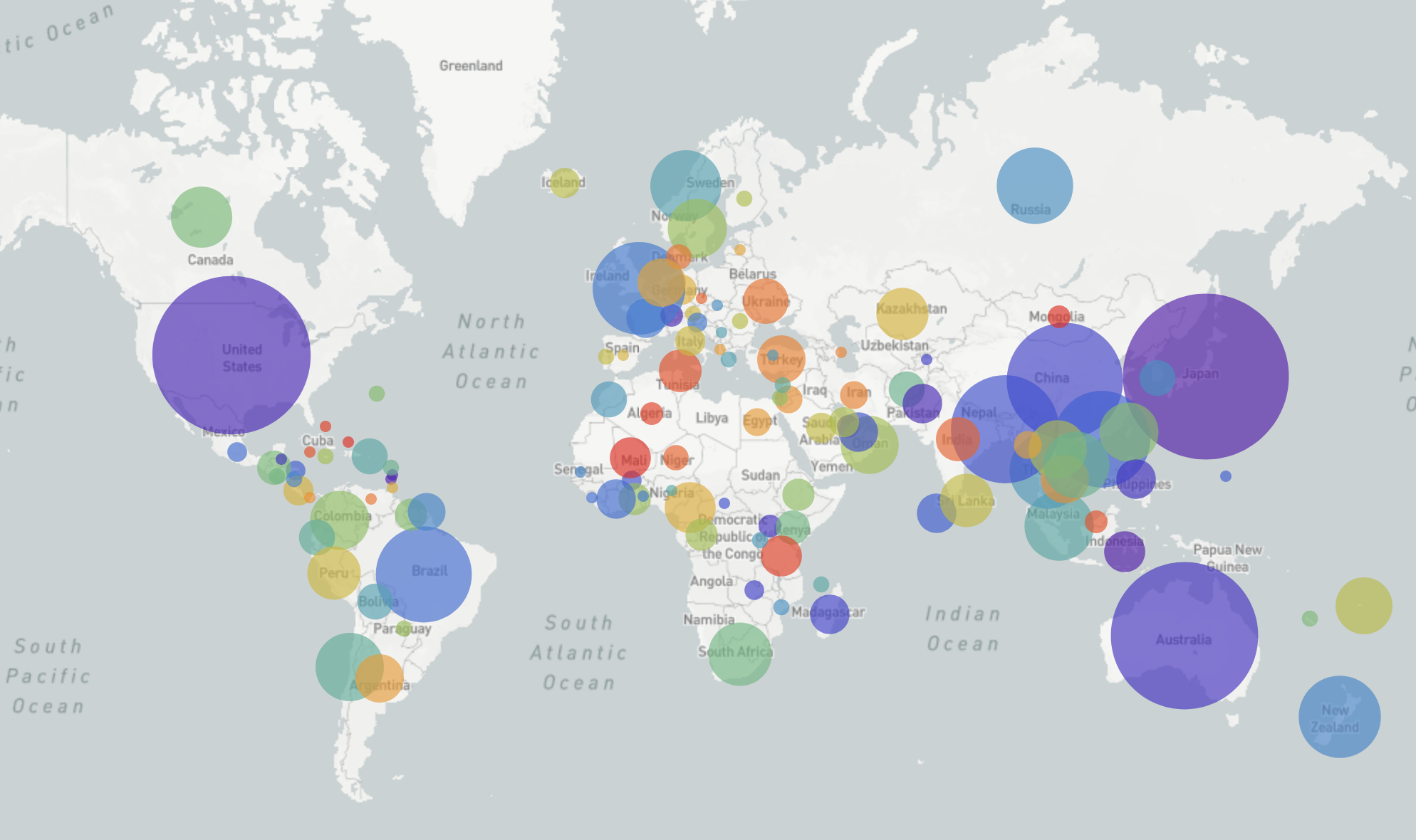
GISRS and GISAID -- Influenza virus surveillance
- comprehensive coverage of the world
- timely sharing of data through GISAID -- often within 2-3 weeks of sampling
- hundreds of sequences per week (in peak months)
→ requires continuous analysis and easy dissemination
→ interpretable and intuitive visualization
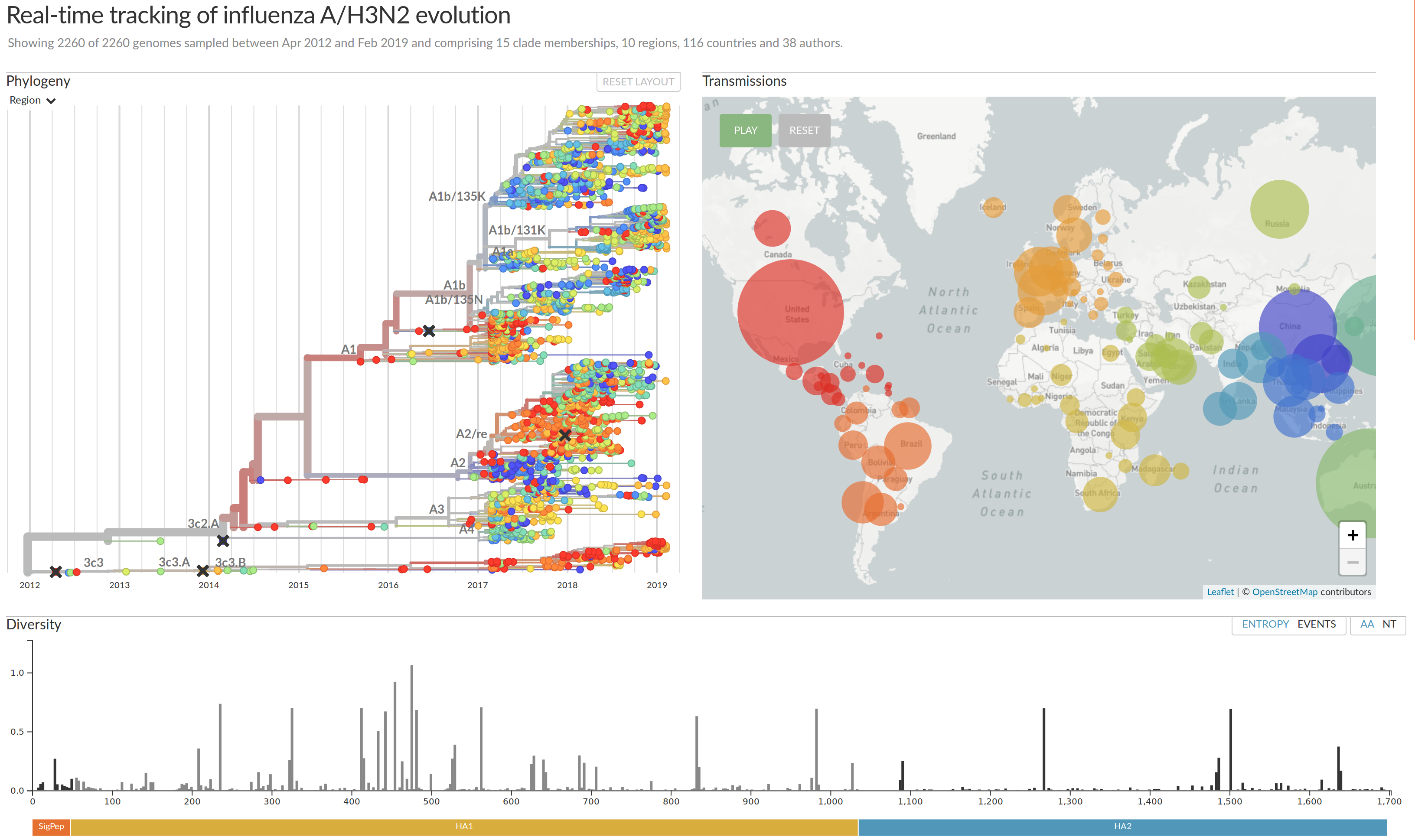
nextflu.org
joint project with Trevor Bedford & his lab
Beyond tracking: can we predict?
Fitness variation in rapidly adapting populations
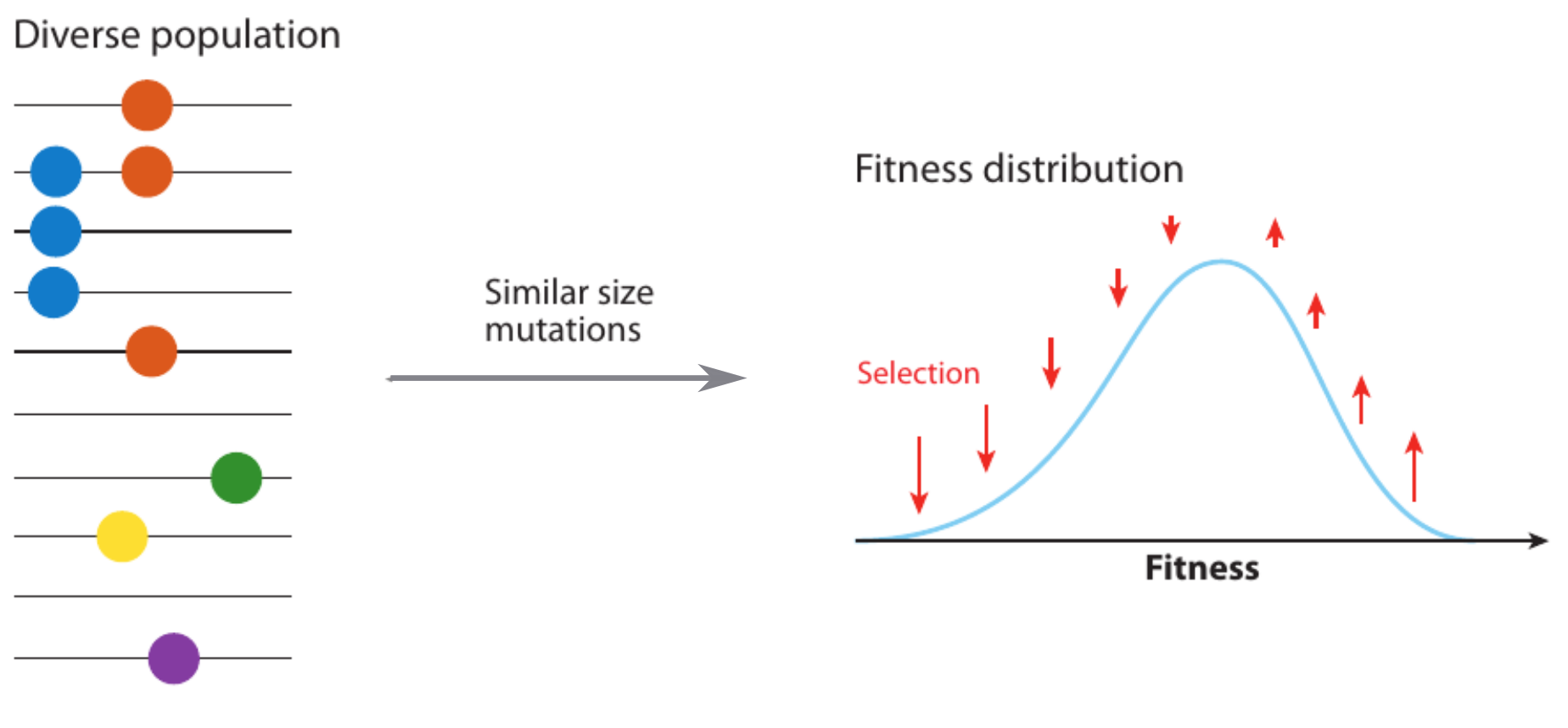
- Speed of adaptation is logarithmic in population size
- Environment (fitness landscape), not mutation supply, determines adaptation
- Different models have universal emerging properties
Neutral/Kingman coalescent
strong selection
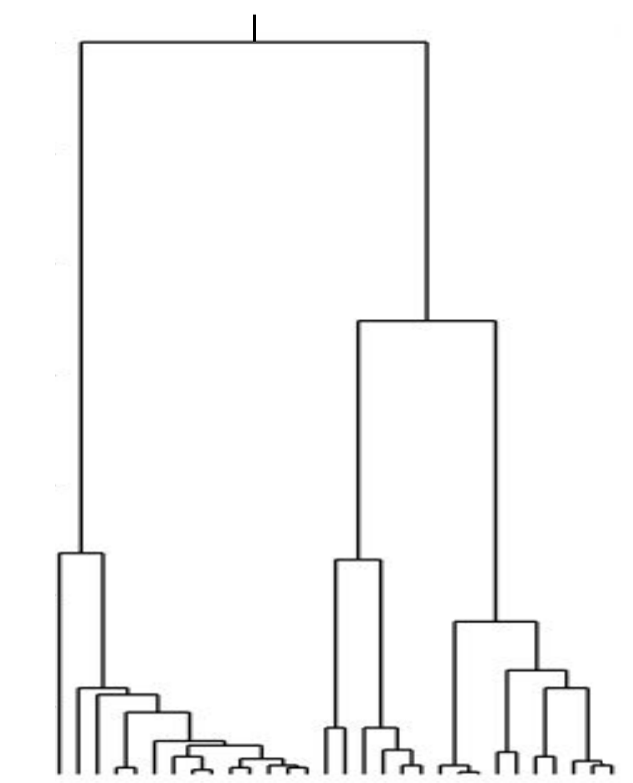

Bolthausen-Sznitman Coalescent
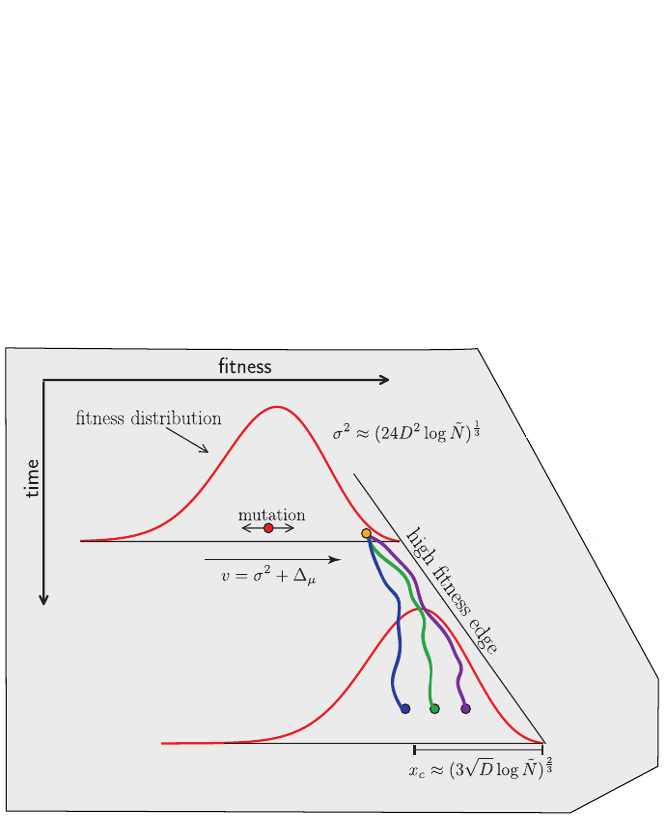
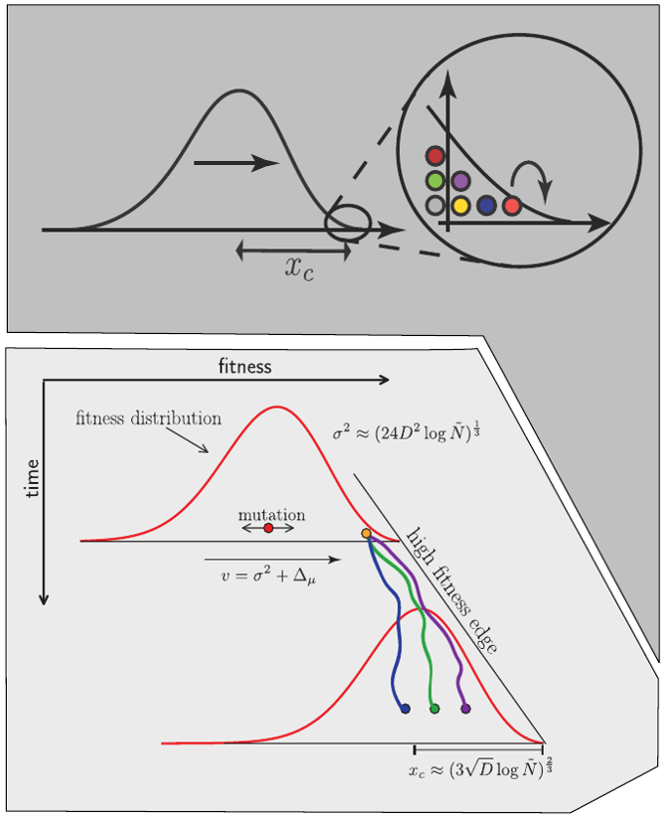
Burst in the tree ↔ high fitness
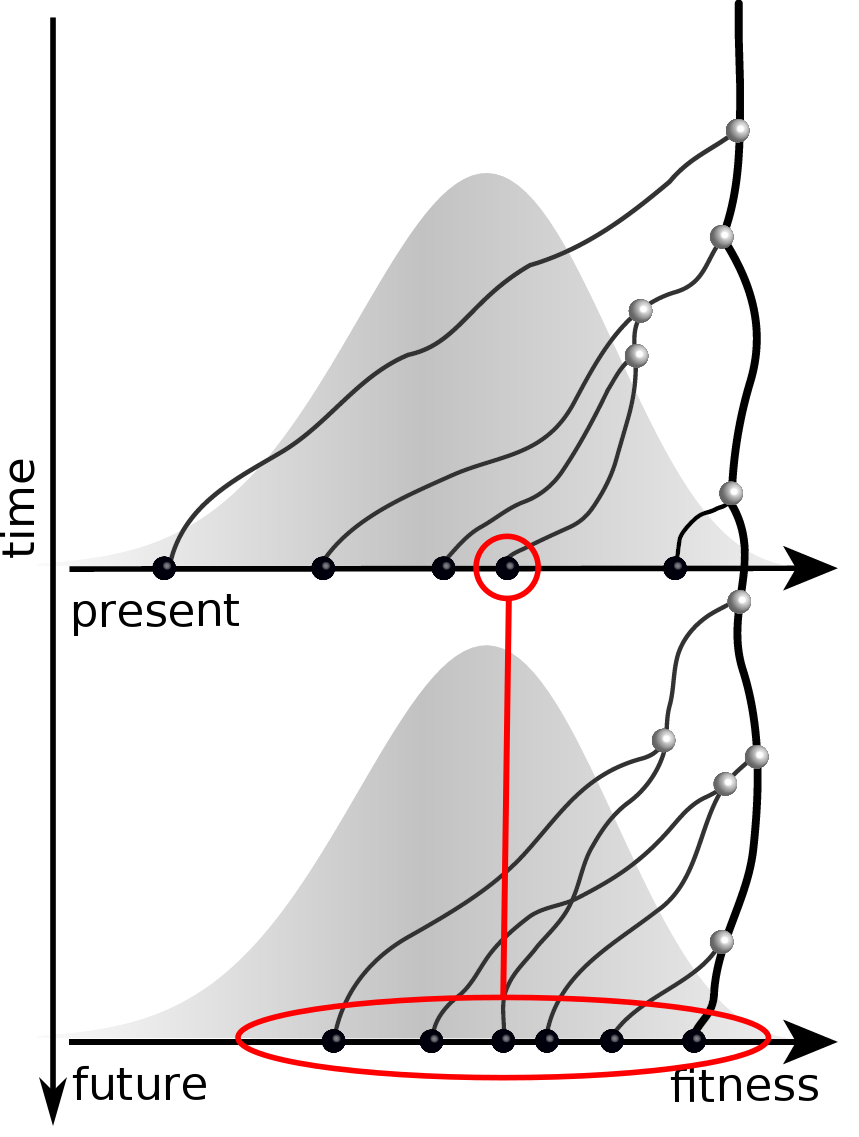
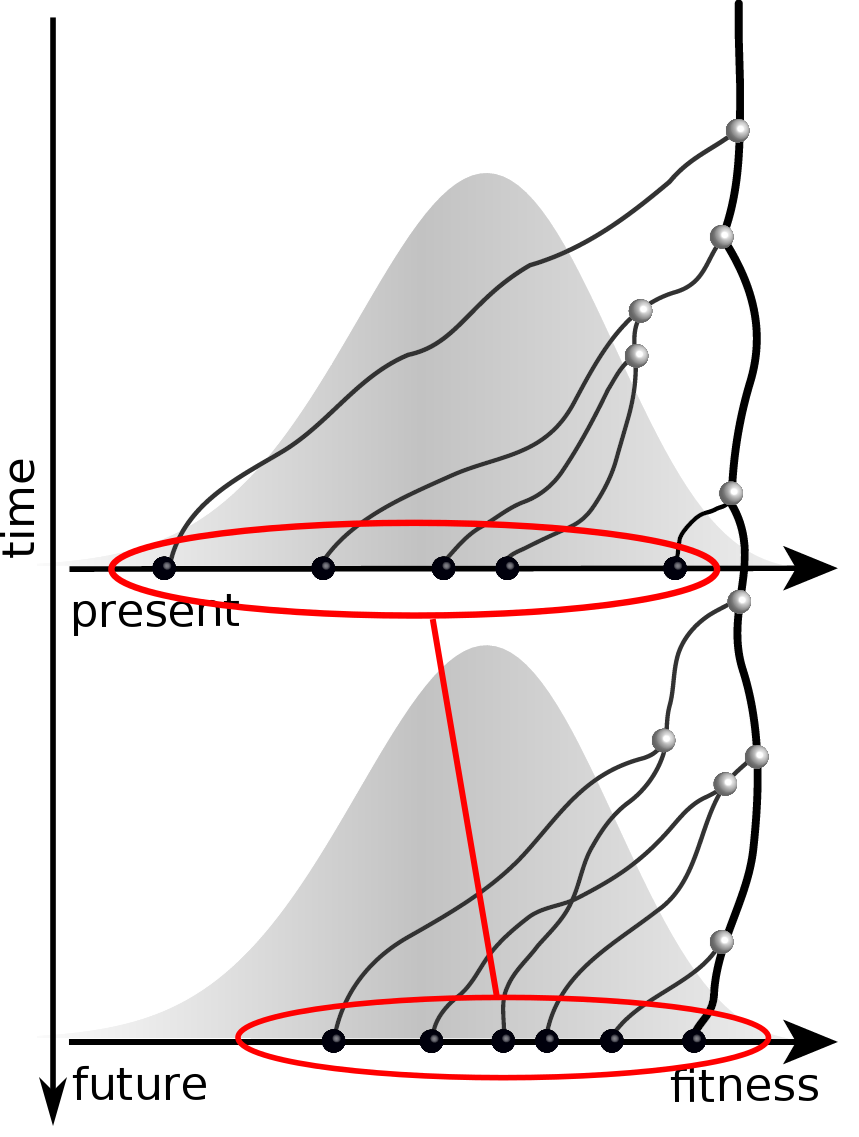
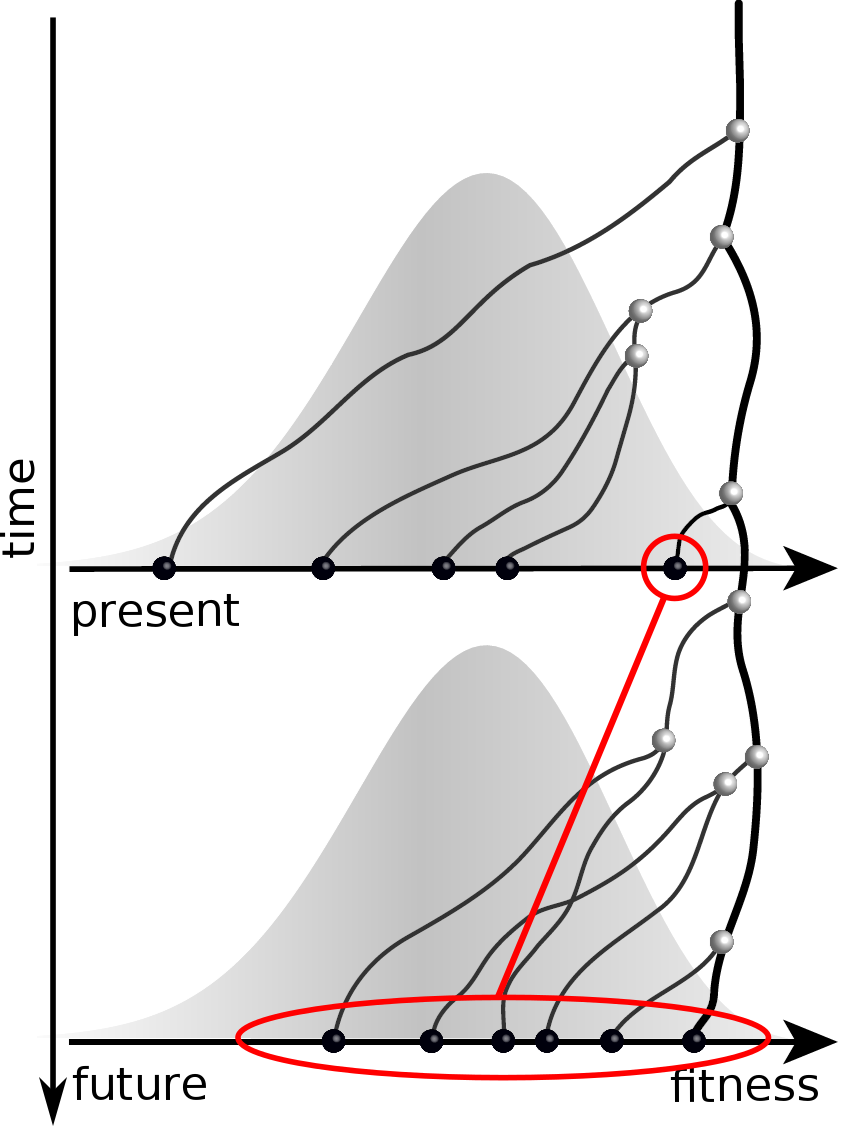
Can we infer fitness from a genomic snapshot?
Fitness inference from trees
$$P(\mathbf{x}|T) = \frac{1}{Z(T)} p_0(x_0) \prod_{i=0}^{n_{int}} g(x_{i_1}, t_{i_1}| x_i, t_i)g(x_{i_2}, t_{i_2}| x_i, t_i)$$
RN, Russell, Shraiman, eLife, 2014
Validation on simulated data
Prediction of the dominating H3N2 influenza strain
- no influenza specific input
- how can the model be improved? (see model by Luksza & Laessig)
Limits of predictability
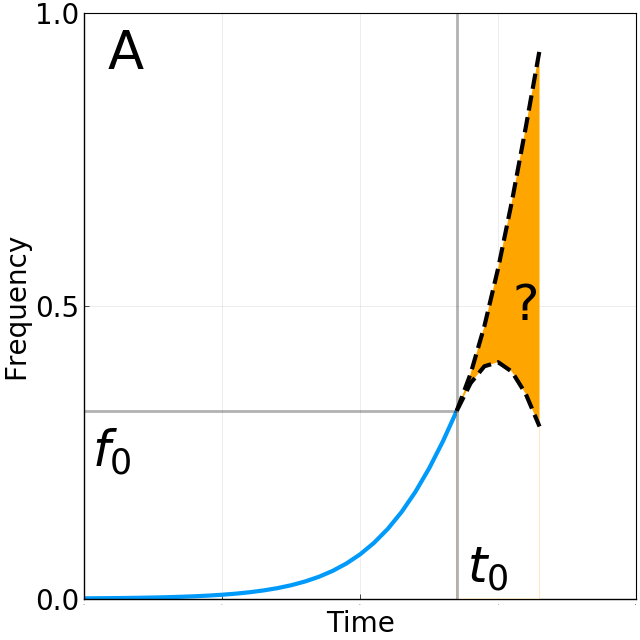
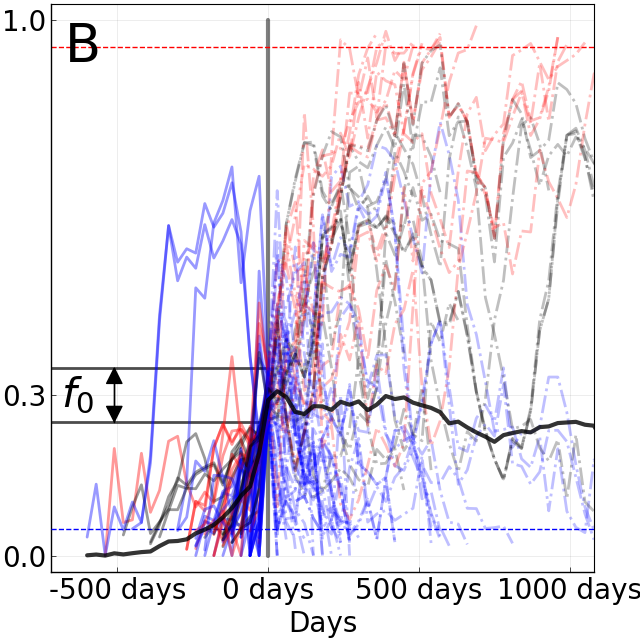
 BBC
BBC
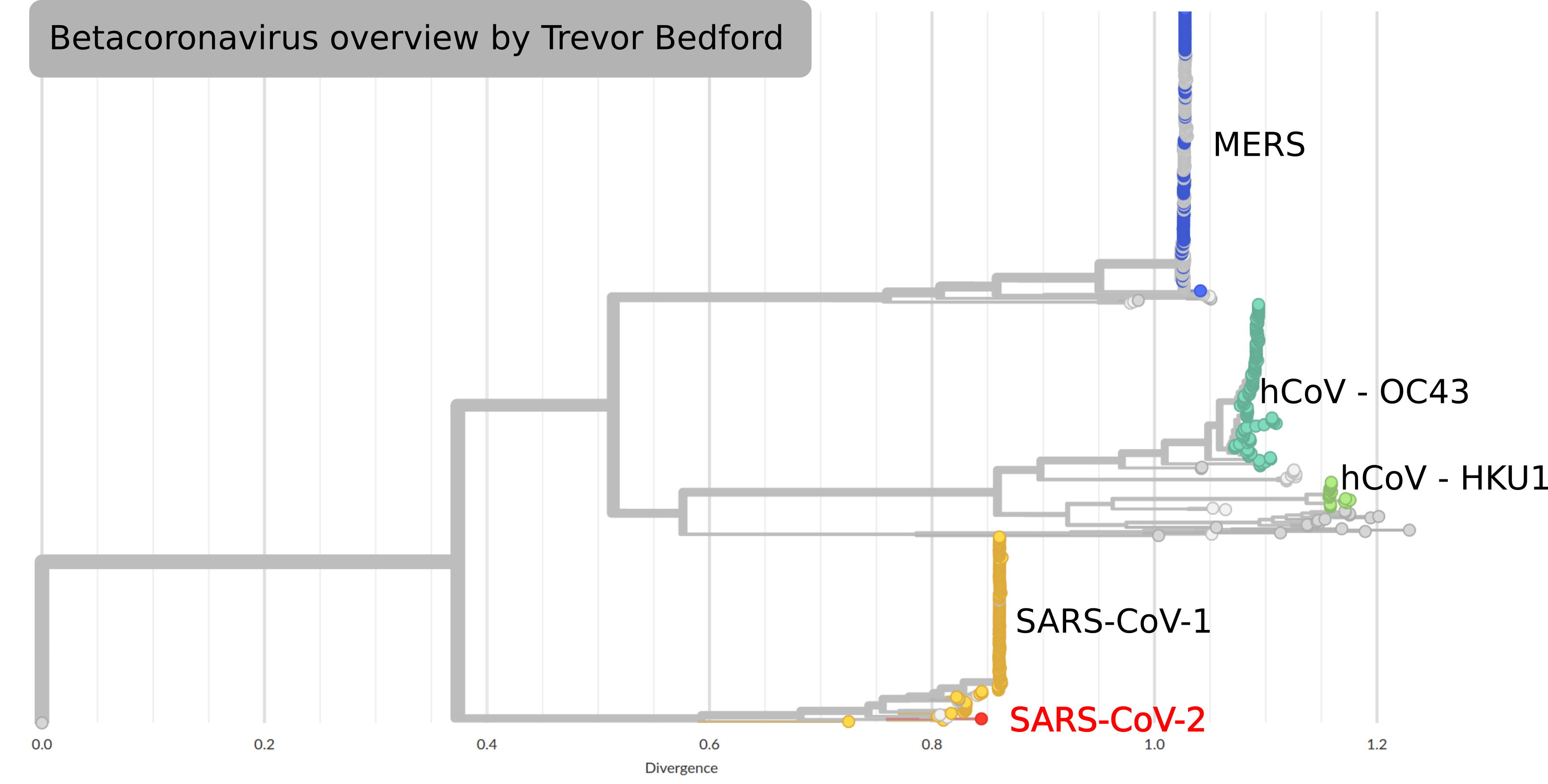 by Trevor Bedford
by Trevor Bedford
 by Trevor Bedford
by Trevor Bedford
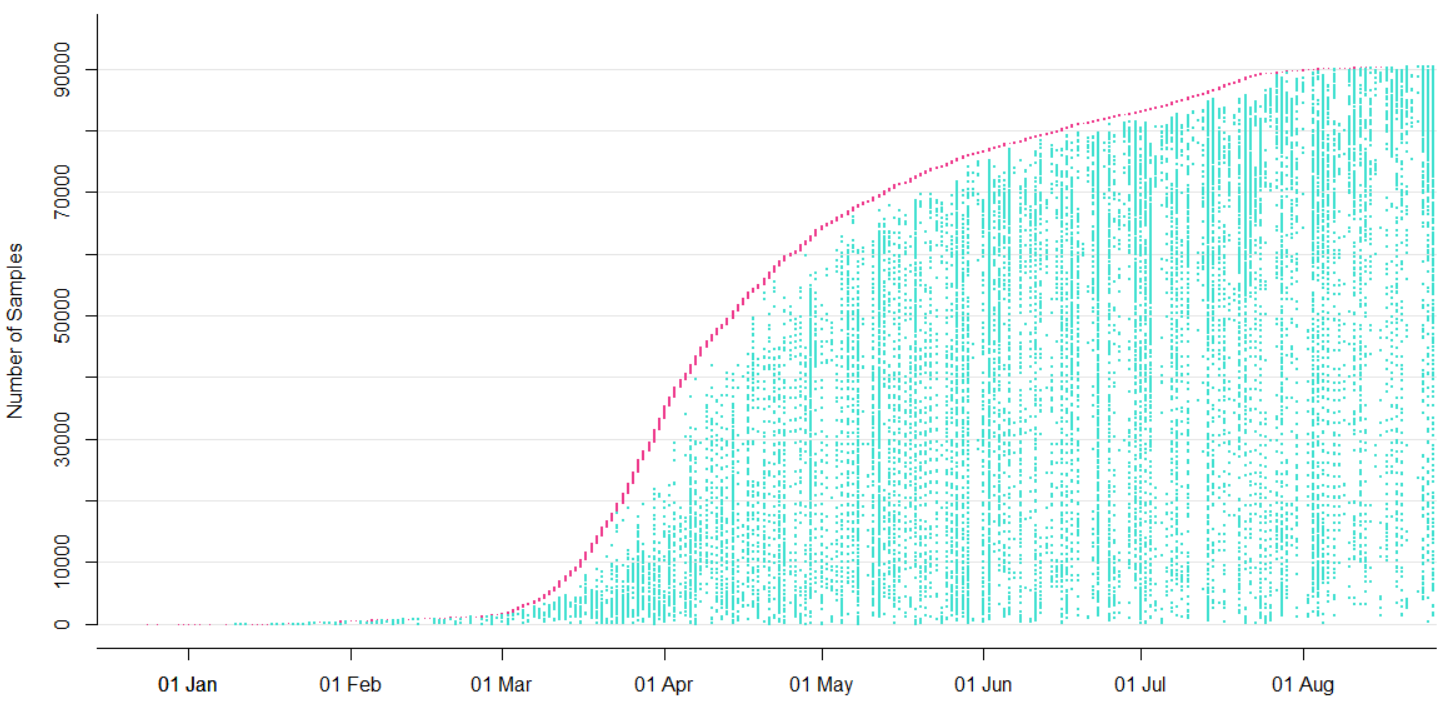
Tracking diversity and spread of SARS-CoV-2 in Nextstrain
Available data on Jan 26
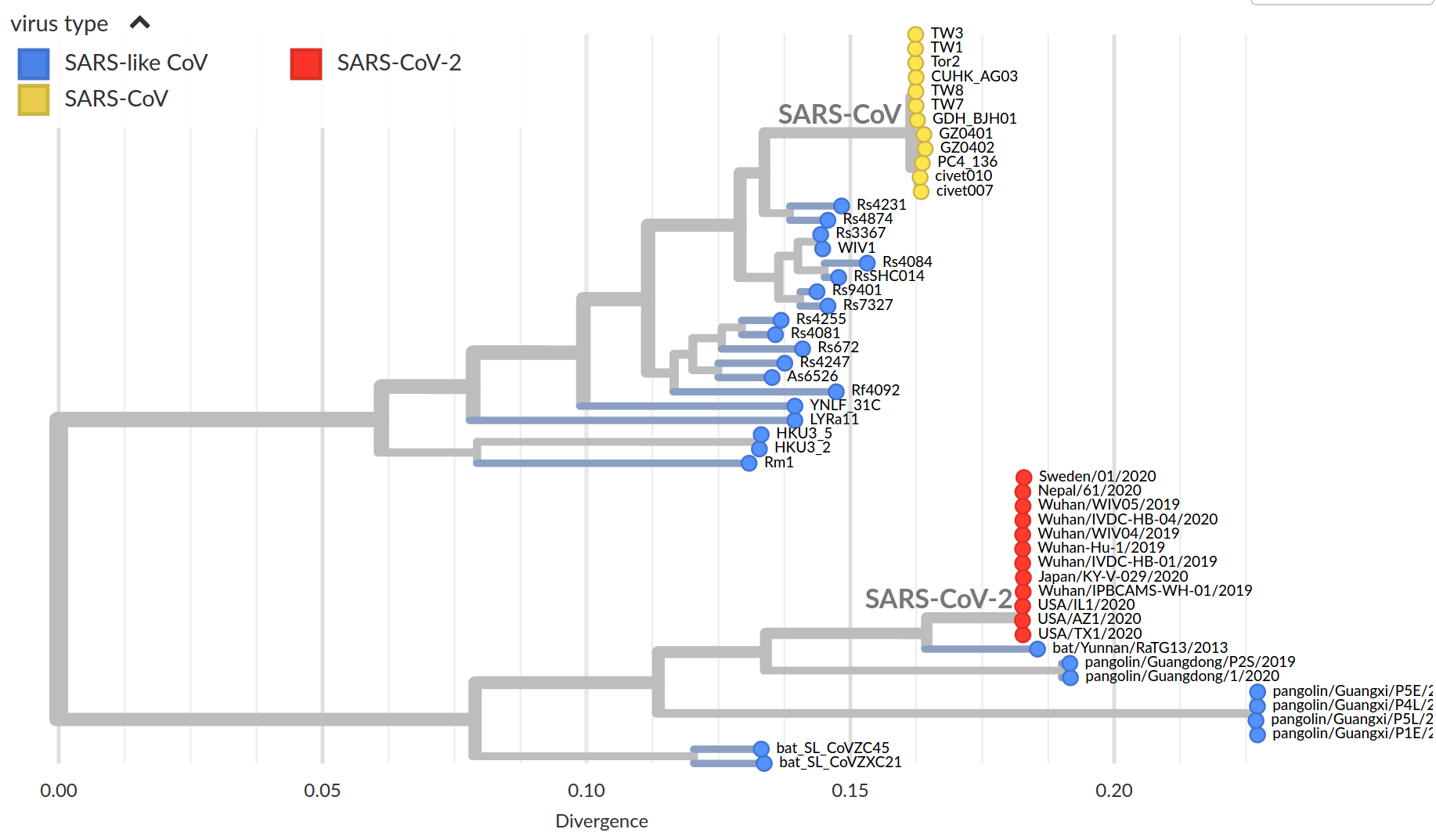
Early genomes differed by only a few mutations, suggesting very recent emergence
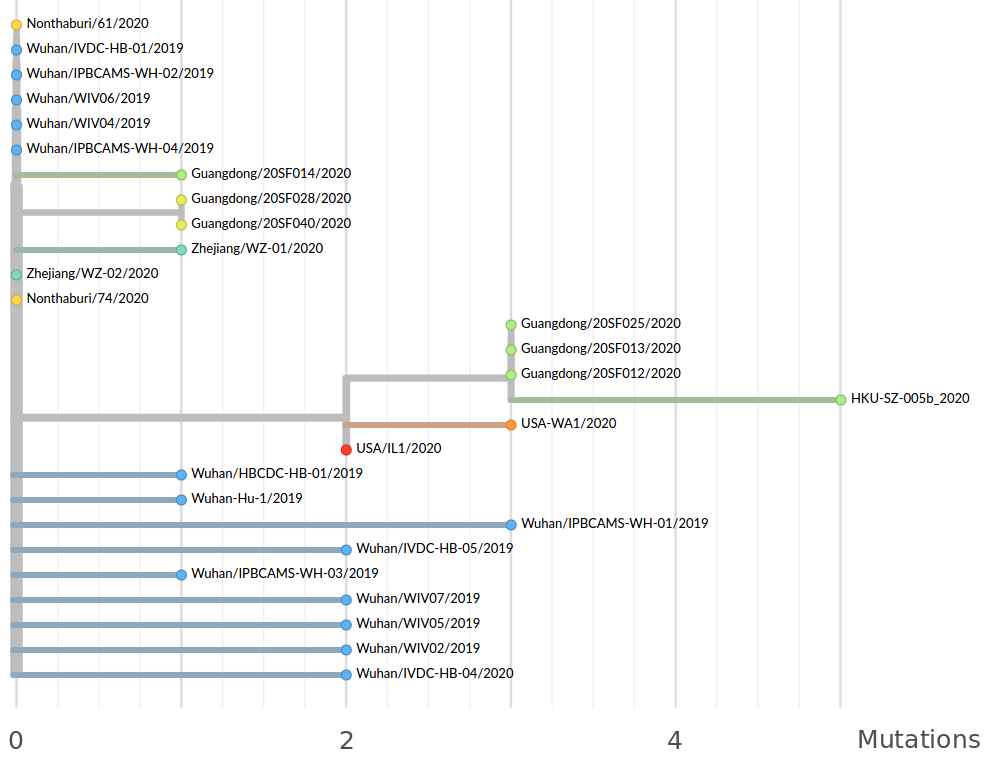
→ the closest to "real-time" we have experienced so far...
Figure by James Hadfield/Emma Hodcroft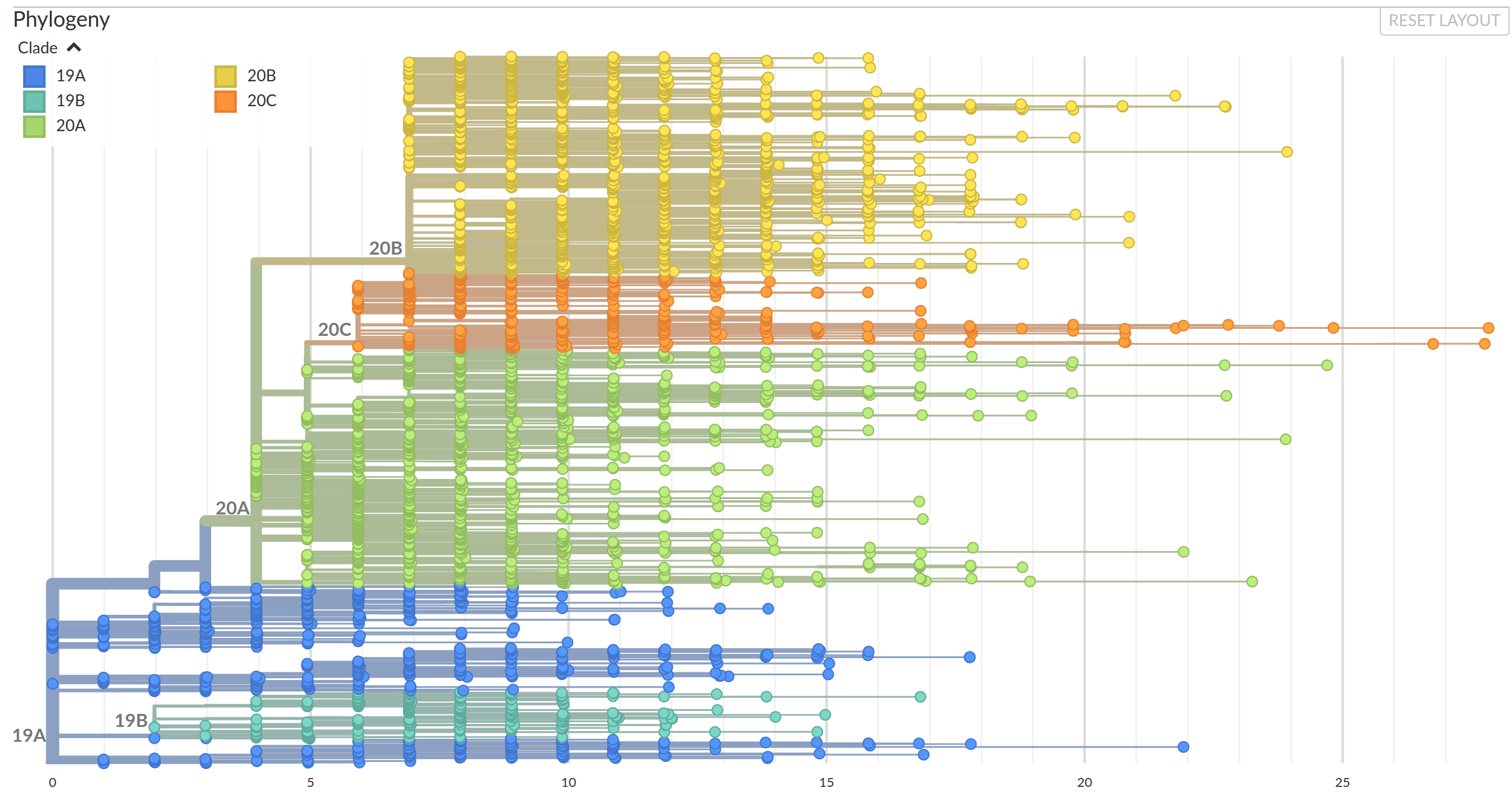
Mutations accumulate constantly, but most of them are irrelevant and rare.
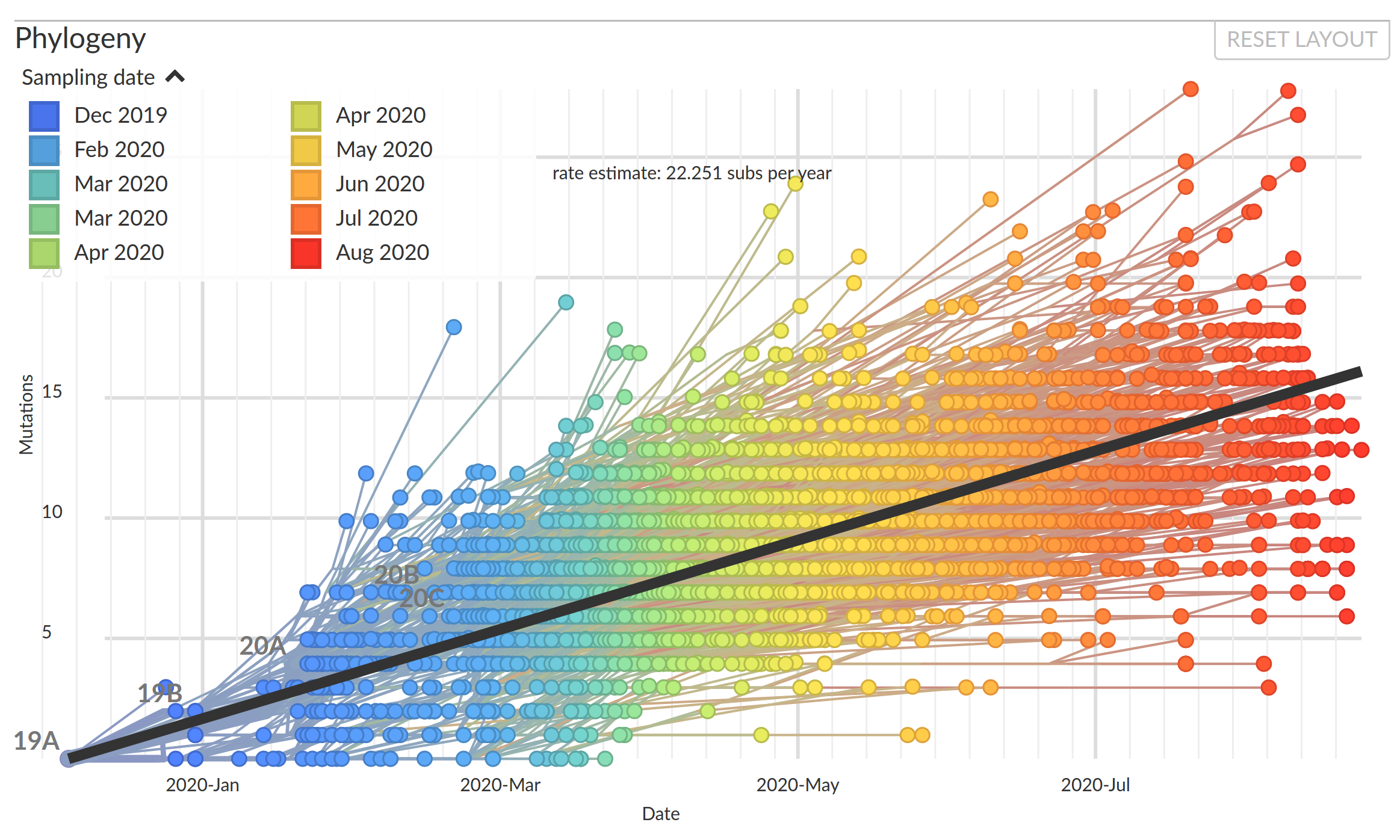
The genome accumulates about two mutations a month...
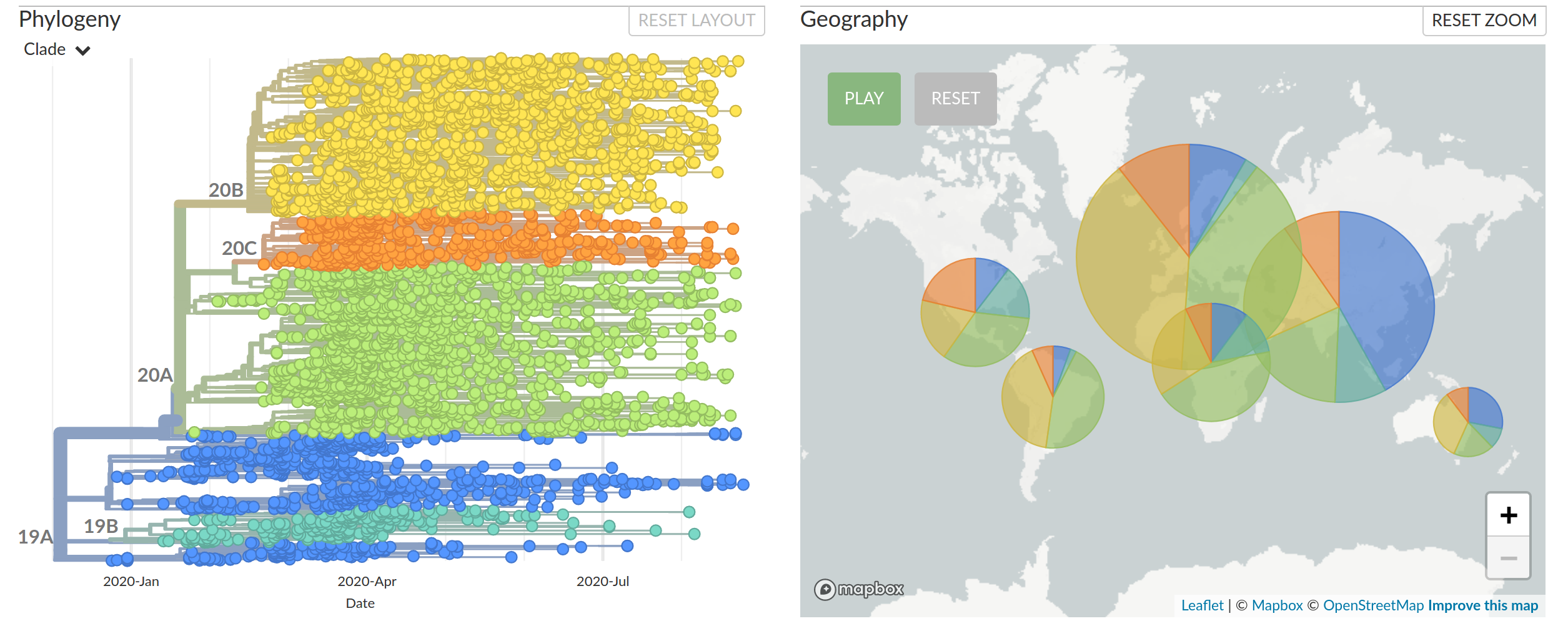
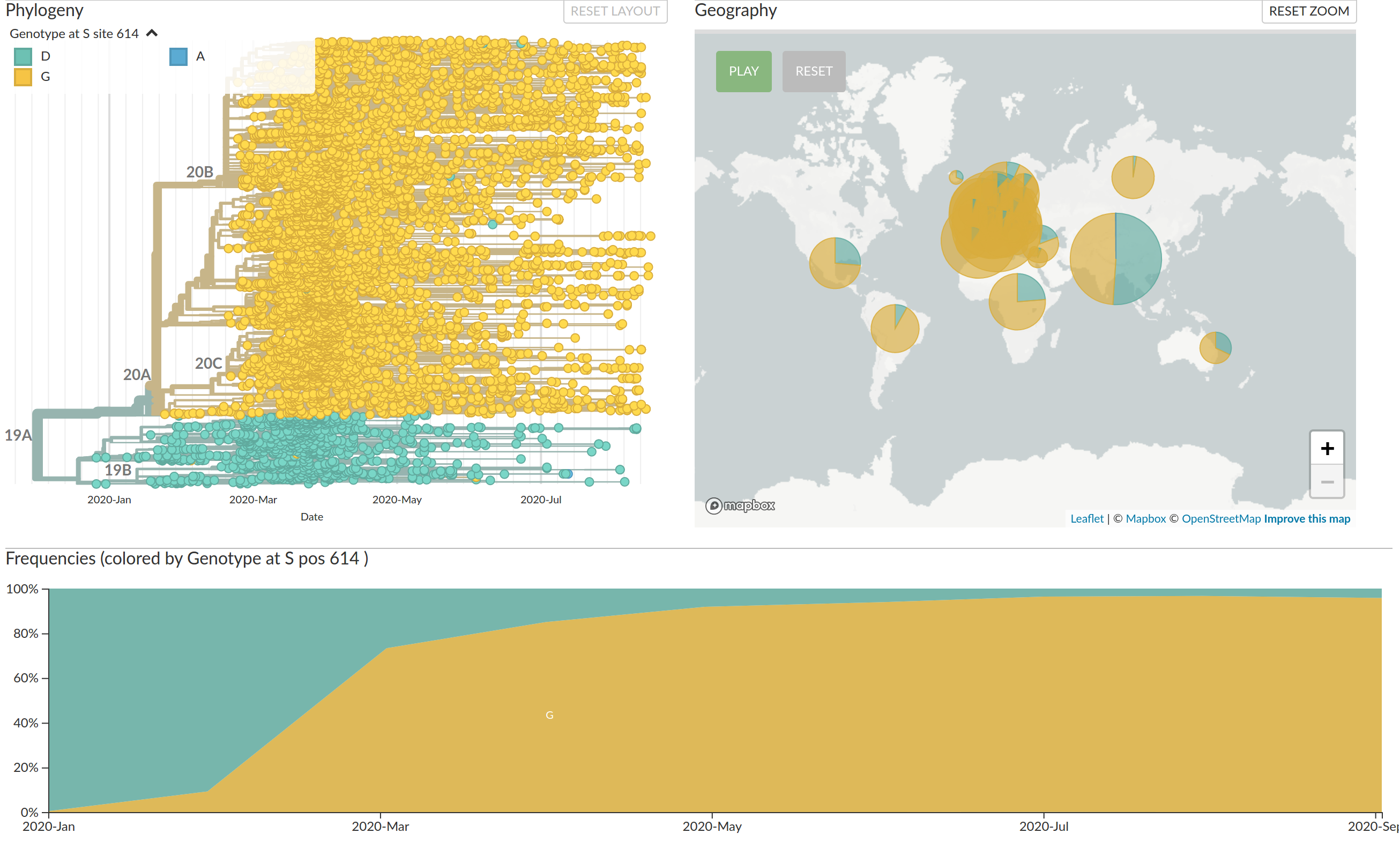
Diversified into multiple global variants. Groups 20A/B/C have taken over.
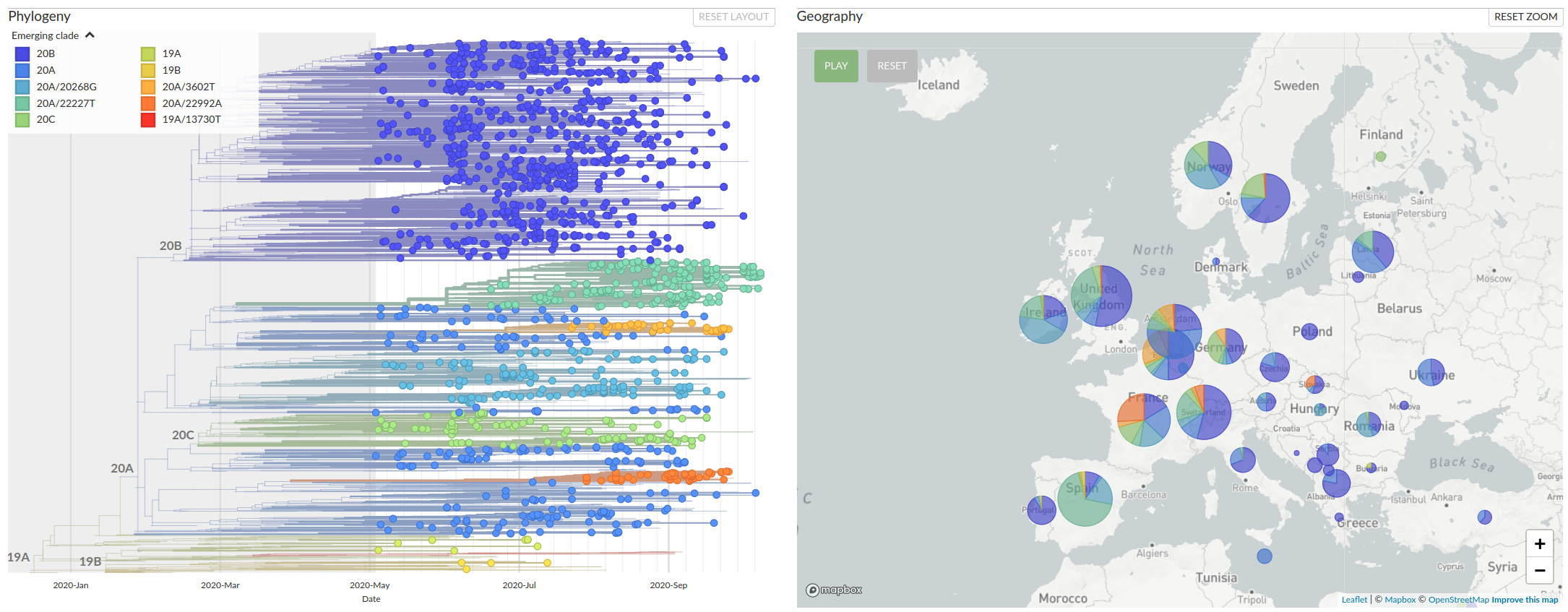
A European clusters in summer 2020
What's next?
Seasonal incidence of influenza viruses

2009 pandemic influenza -- US
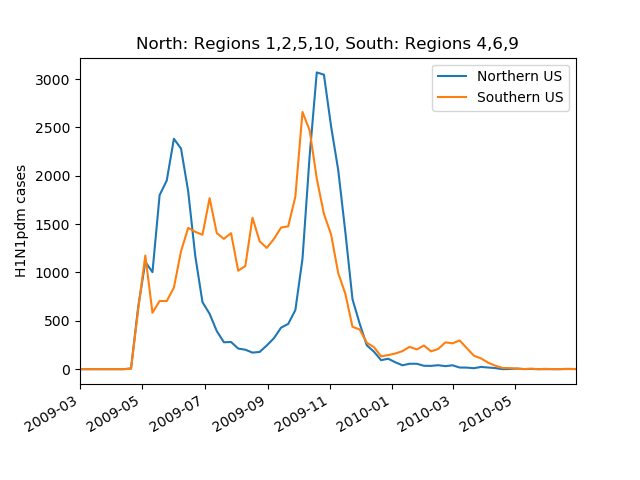
 by Trevor Bedford
by Trevor Bedford
Human corona viruses have pronounced seasonal prevalence (Sweden)
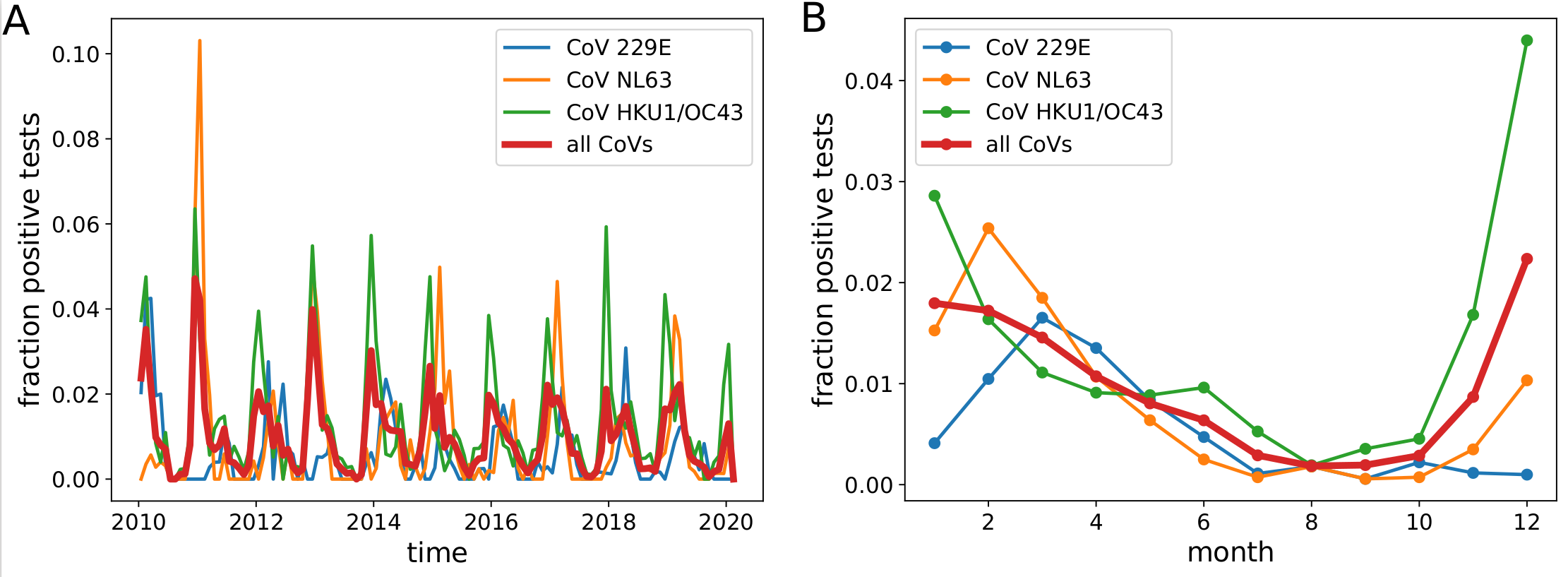
- Most respiratory virus including established CoVs show seasonality
- Little direct evidence; absolute effect of seasonality unknown
- But expect control of the virus to be harder in winter
Potential transition to an endemic seasonal virus
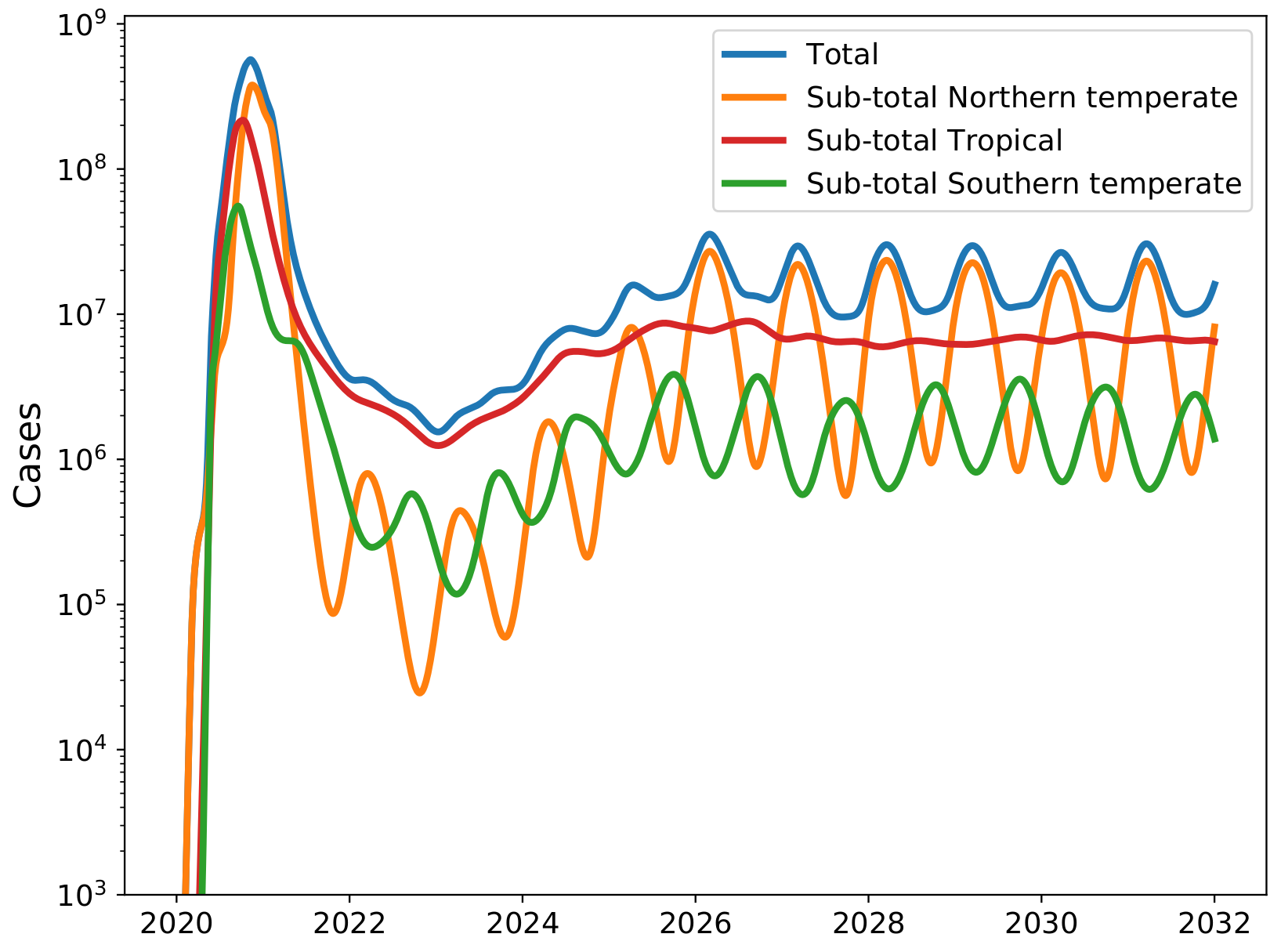
Influenza and Theory acknowledgments





- Boris Shraiman
- Colin Russell
- Trevor Bedford
- Pierre Barrat
- Oskar Hallatschek
- All the NICs and WHO CCs that provide influenza sequence data
- The WHO CCs in London and Atlanta for providing titer data



Acknowledgments
Trevor Bedford and his lab -- terrific collaboration since 2014

especially James Hadfield, Emma Hodcroft, Ivan Aksamentov, Moira Zuber, and Tom Sibley
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID


Acknowledgments
- Robert Dyrdak
- Jan Albert
- Valentin Druelle
- Emma Hodcroft
