Evolution and spread of Influenza, HIV, and SARS-CoV-2
Richard Neher
Biozentrum, University of Basel
Human seasonal influenza viruses

Positive tests for influenza in the USA by week
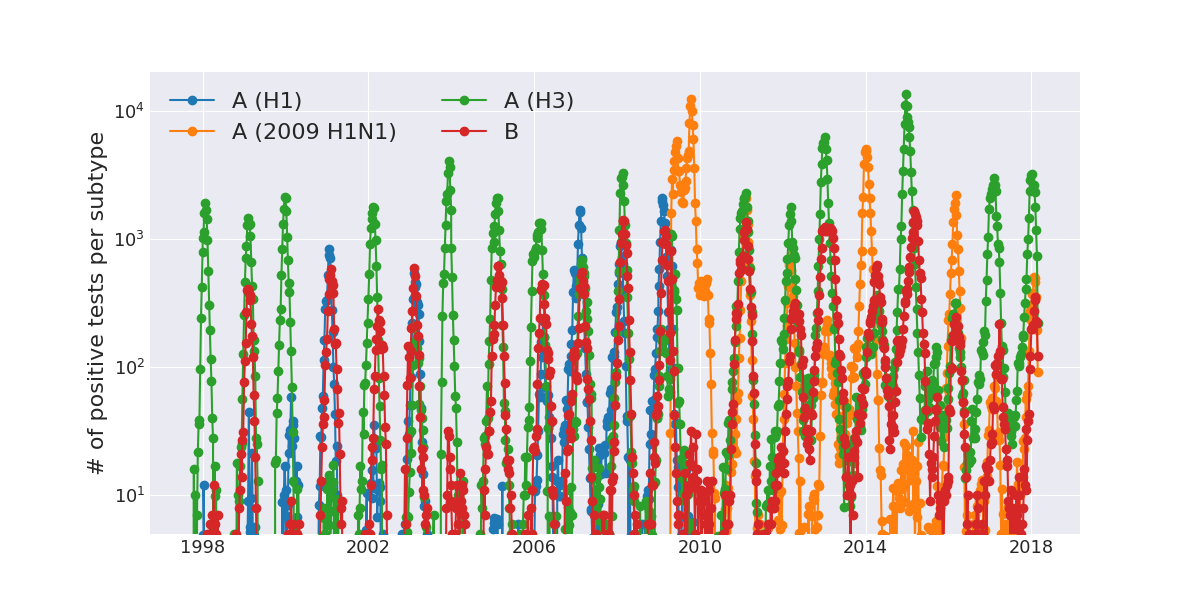 Data by the US CDC
Data by the US CDC
Virus genomes change rapidly through time
A/Brisbane/100/2014
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
A/Brisbane/1000/2015
GGATAATTCTATTAACCATGAAGACTATTATTGCTTT...
A/Brisbane/1/2017
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
... hundreds of thousands of sequences...
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
A/Brisbane/1000/2015
GGATAATTCTATTAACCATGAAGACTATTATTGCTTT...
A/Brisbane/1/2017
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
... hundreds of thousands of sequences...
Genome sequences record the spread of pathogens




in SARS-CoV-2 a new mutation accumulates about every 2 weeks



- Influenza viruses evolve to avoid human immunity
- Vaccines need frequent updates

Where does all that evolution happen?
Suppressive therapy and the latent reservoir:
Where is HIV hiding?
The latent proviral DNA reservoir
- Viruses integrate but don't express → pro-viral DNA reservoir
- Therapy fully suppresses virus replication, but interruption results in rebound
- When is the reservoir established? How is it maintained?
p17 sequencing from PBMCs on fully suppressive therapy
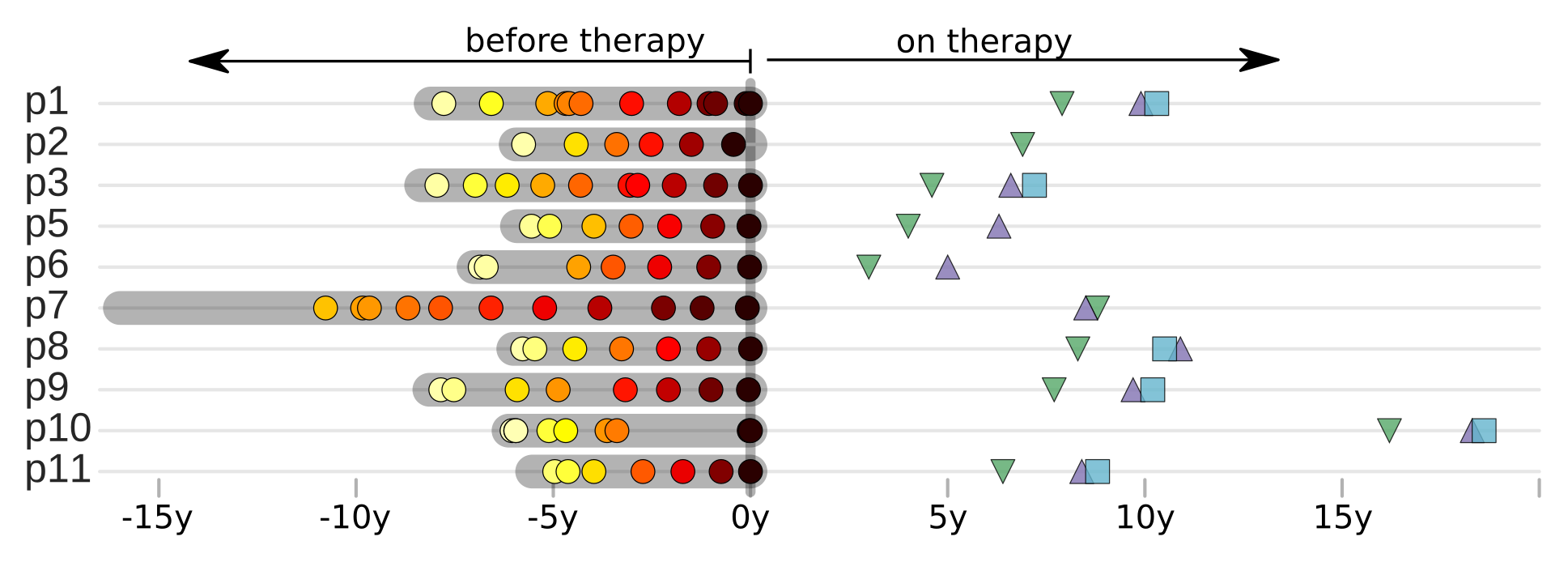 Brodin et al, eLife, 2016
Brodin et al, eLife, 2016
Proviral DNA mostly identical to pre-therapy RNA
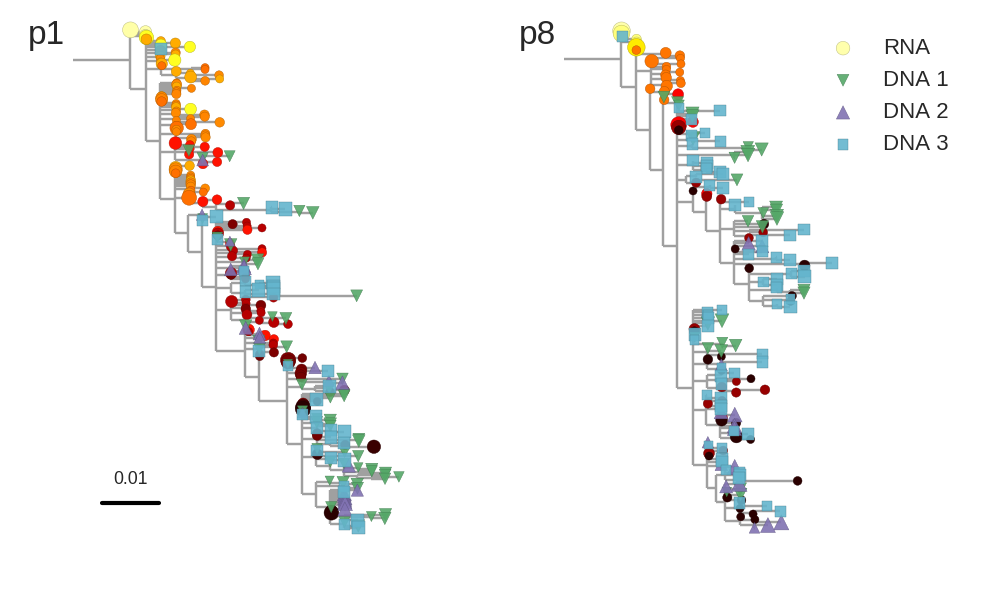
No evidence of ongoing evolution
Most proviral DNA originates from shortly after start of therapy
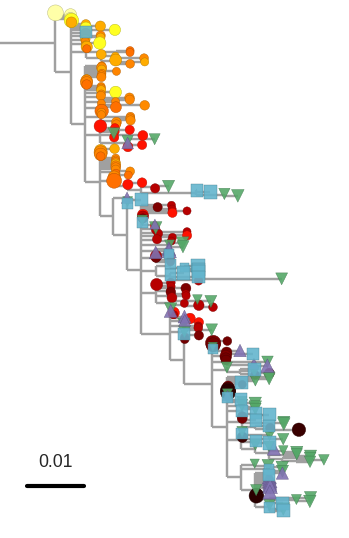
- latent HIV → barcode of a T-cell lineage
- all latent integrated virus derives from late infection
Real-time tracking of SARS-CoV-2
- hundreds of new sequences every day
- more than 200k sequences right now
- comprehensive analysis require hours to days to complete
→ requires continuous analysis and easy dissemination
→ interpretable and intuitive visualization
nextstrain.org
joint project with Trevor Bedford & his lab
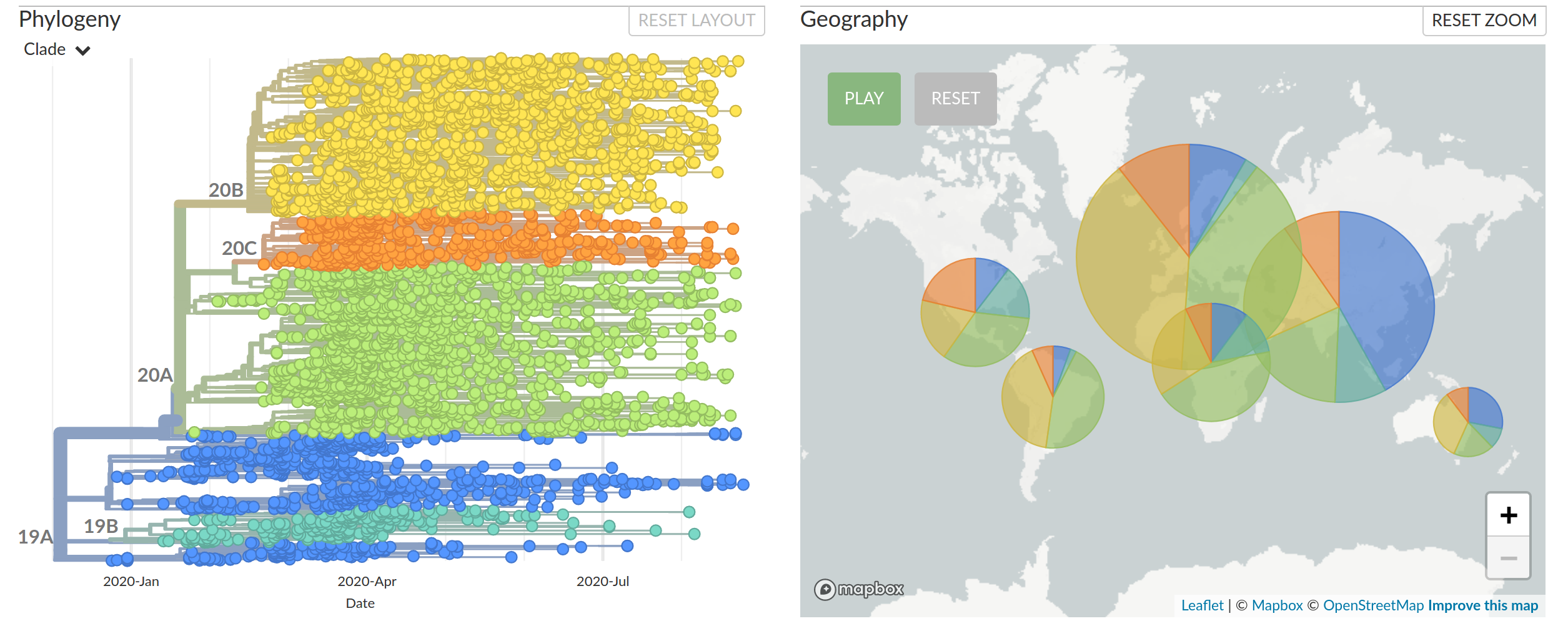
Diversified into multiple global variants. Groups 20A/B/C have taken over.
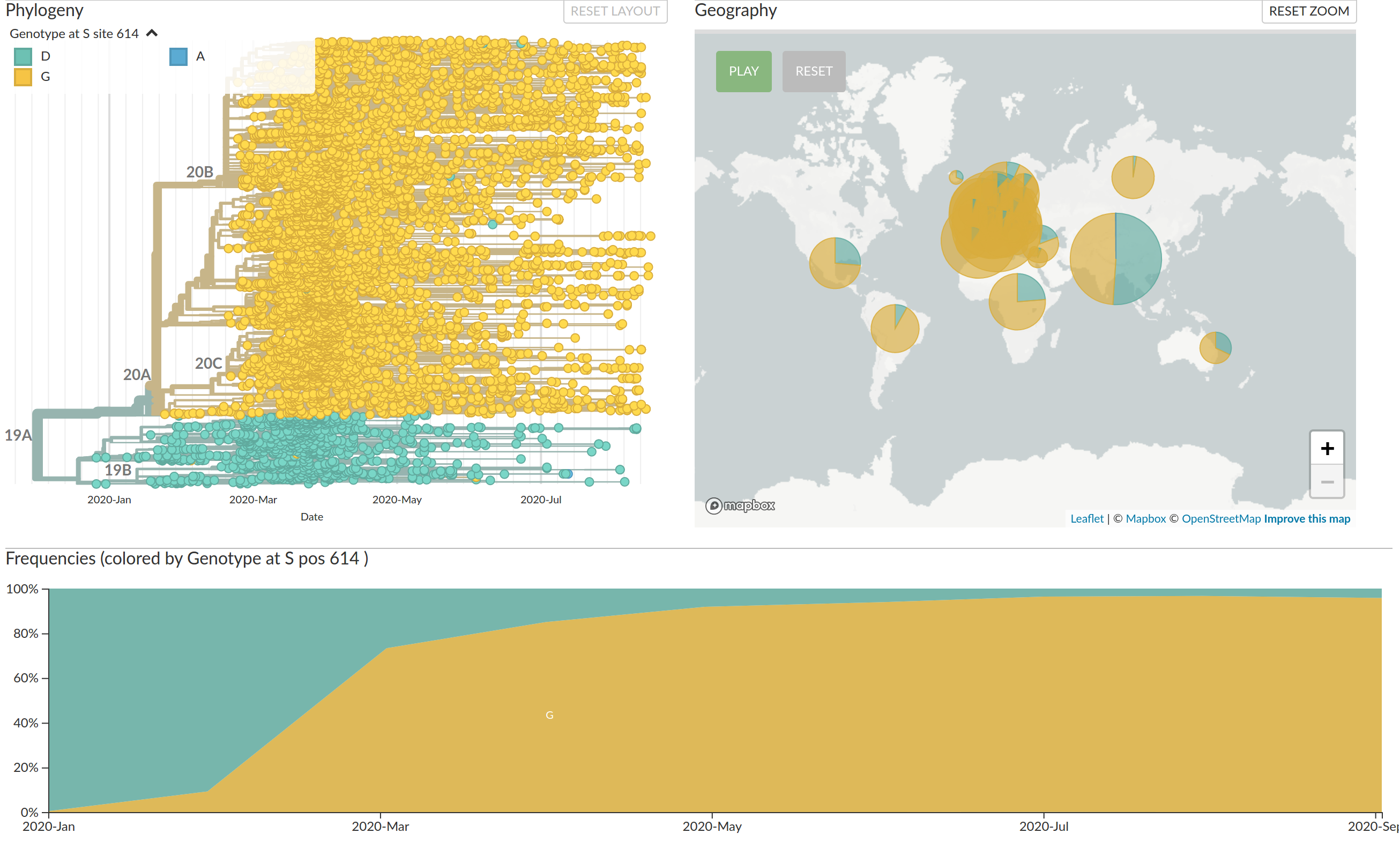
Large scale spread of the virus
Regional variants emerged over the summer
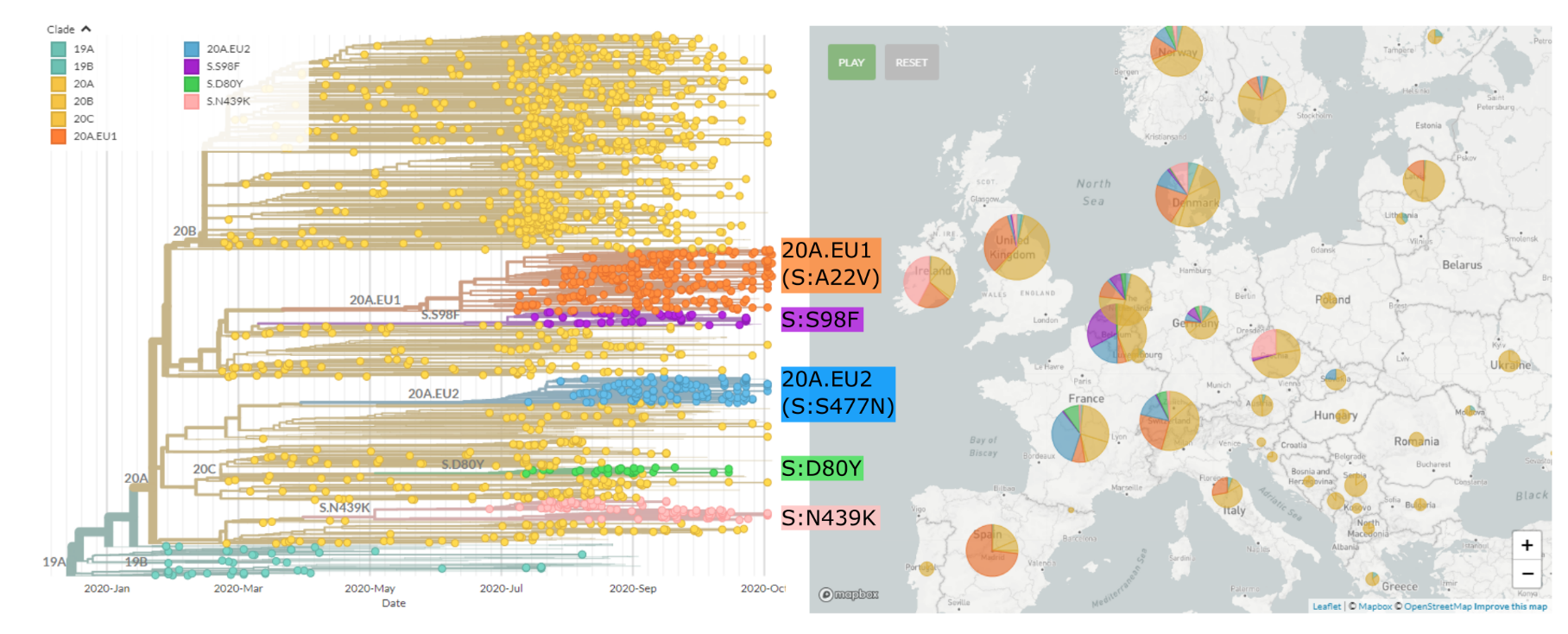
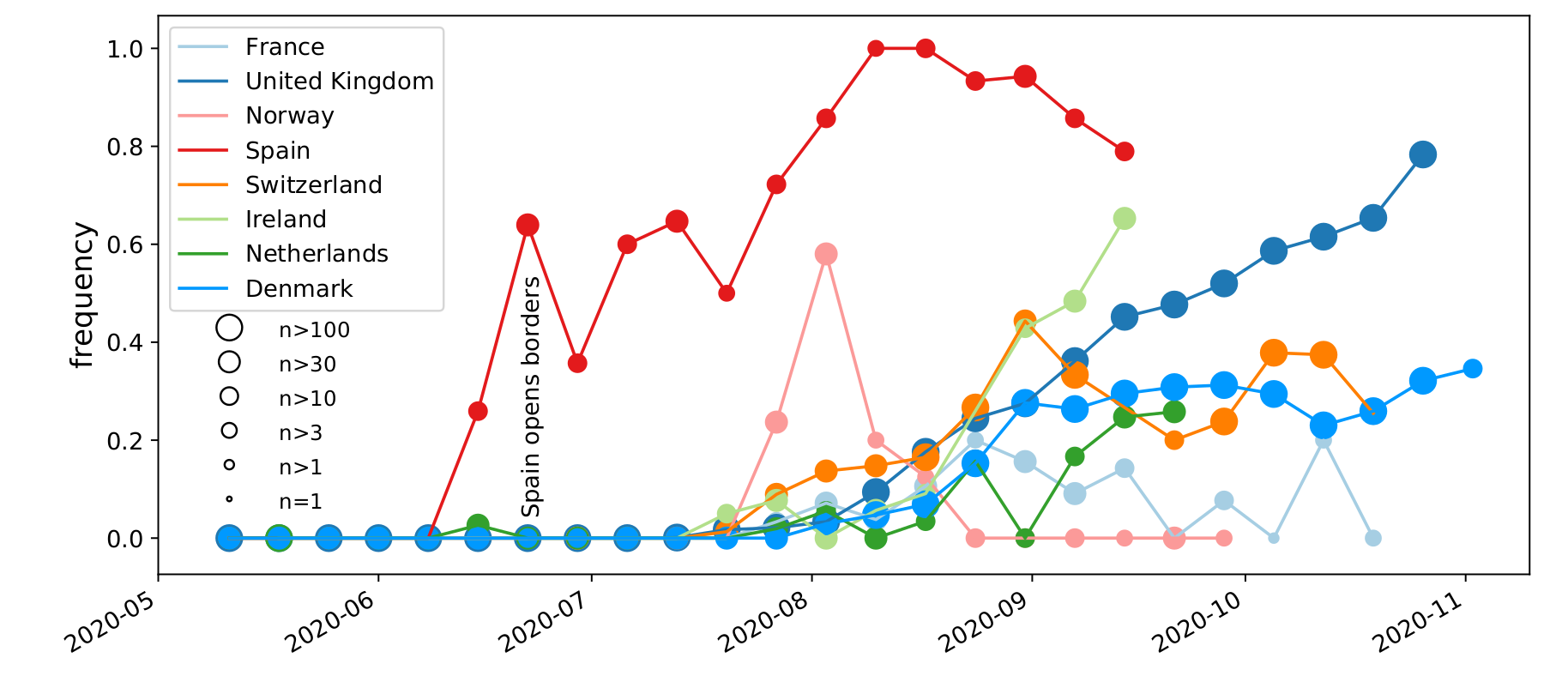
Spread of a variant through travel and tourism
Acknowledgments
- Fabio Zanini
- Jan Albert
- Johanna Brodin
- Christa Lanz
- Göran Bratt
- Lina Thebo
- Vadim Puller



Acknowledgments
- Emma Hodcroft
- Moira Zuber
- Inaki Comas
- Fernando Gonzales
- Tanja Stadler
- Sarah Nadeau
- Tim Vaughan
- Jesse Bloom
- David Veesler
