Tracking and predicting the evolution RNA viruses
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/201702_compare.html
RNA viruses are primary pandemic threats
- influenza virus (spanish flu 1918, "swine flu" 2009, H5N1, ...)
- SARS (Severe Acute Respiratory Syndrome, coronavirus)
- MERS (Middle East respiratory syndrome, coronavirus)
- Ebola (filovirus)
- Zika virus (flavivirus)
- ...
Human seasonal influenza viruses

Surveillance of human seasonal influenza viruses
- WHO CCs and NICs sequence and phenotype 100s of viruses per month
- Sequences allow us to track how the virus is spreading and changing







- Influenza virus evolves to avoid human immunity
- Vaccines need frequent updates

nextflu.org
joint work with Trevor Bedford & his lab
nextstrain.org
joint work with Trevor Bedford & his lab
NextStrain architecture
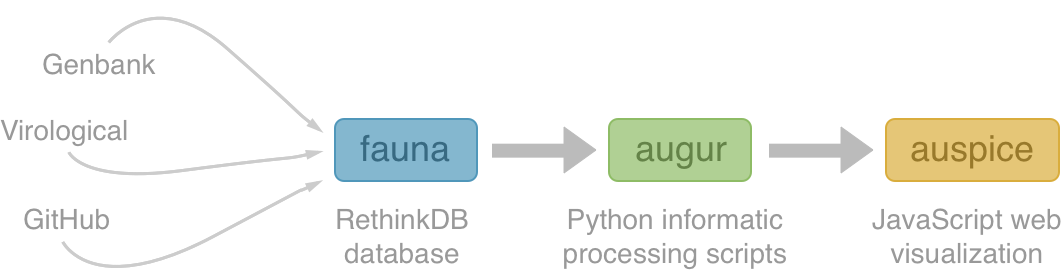
Using treetime to rapidly compute timetrees
Modeling and predicting influenza evolution
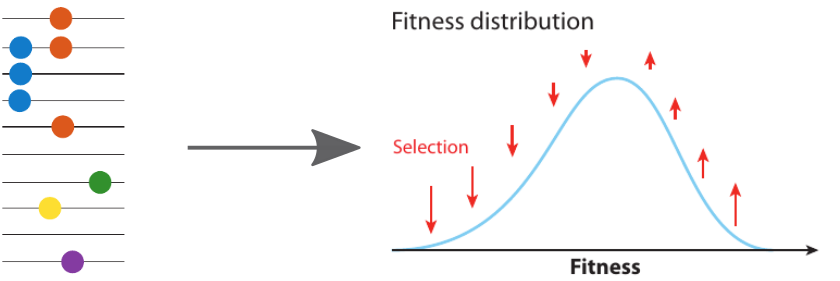
Predicting evolution

Given the branching pattern,
RN, Russell, Shraiman, eLife, 2014
- can we predict fitness?
- pick the closest relative of the future?
Fitness inference from trees
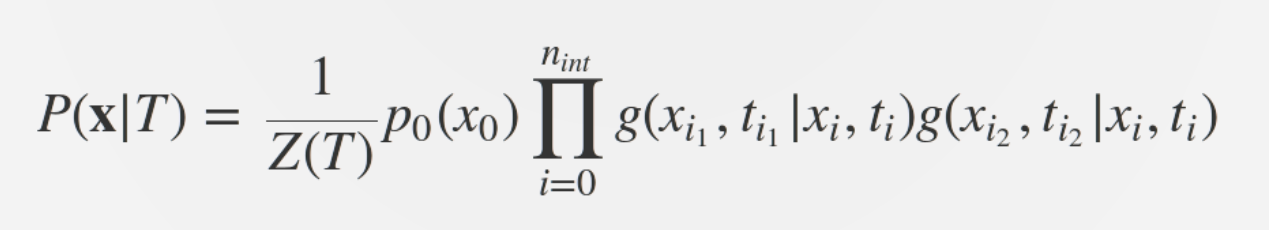
Prediction of the dominating H3N2 influenza strain
RN, Russell, Shraiman, eLife, 2014Virus evolution takes place within the host
Deep longitudinal sampling is necessary to monitor evolution in detail
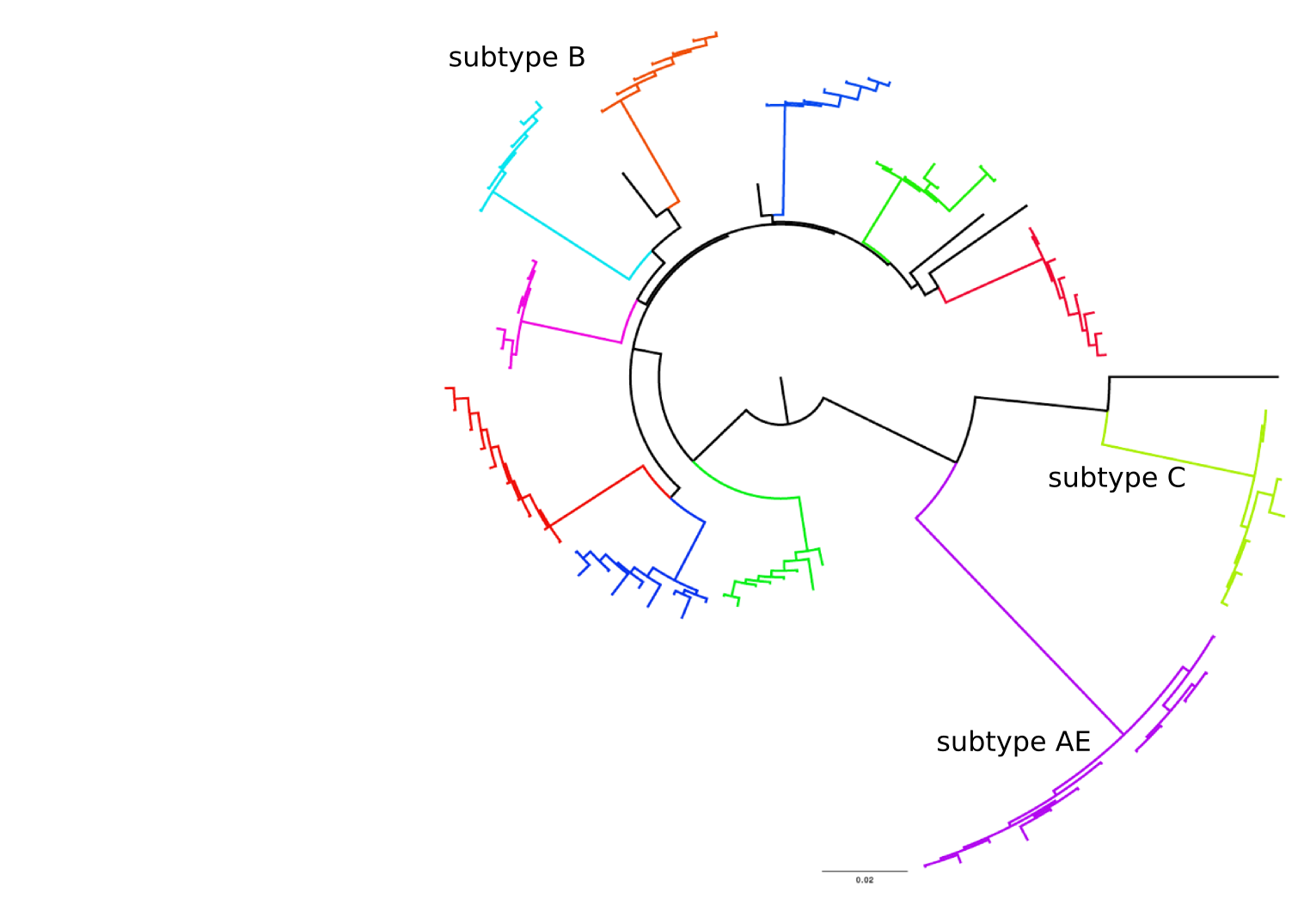
Evolution of HIV
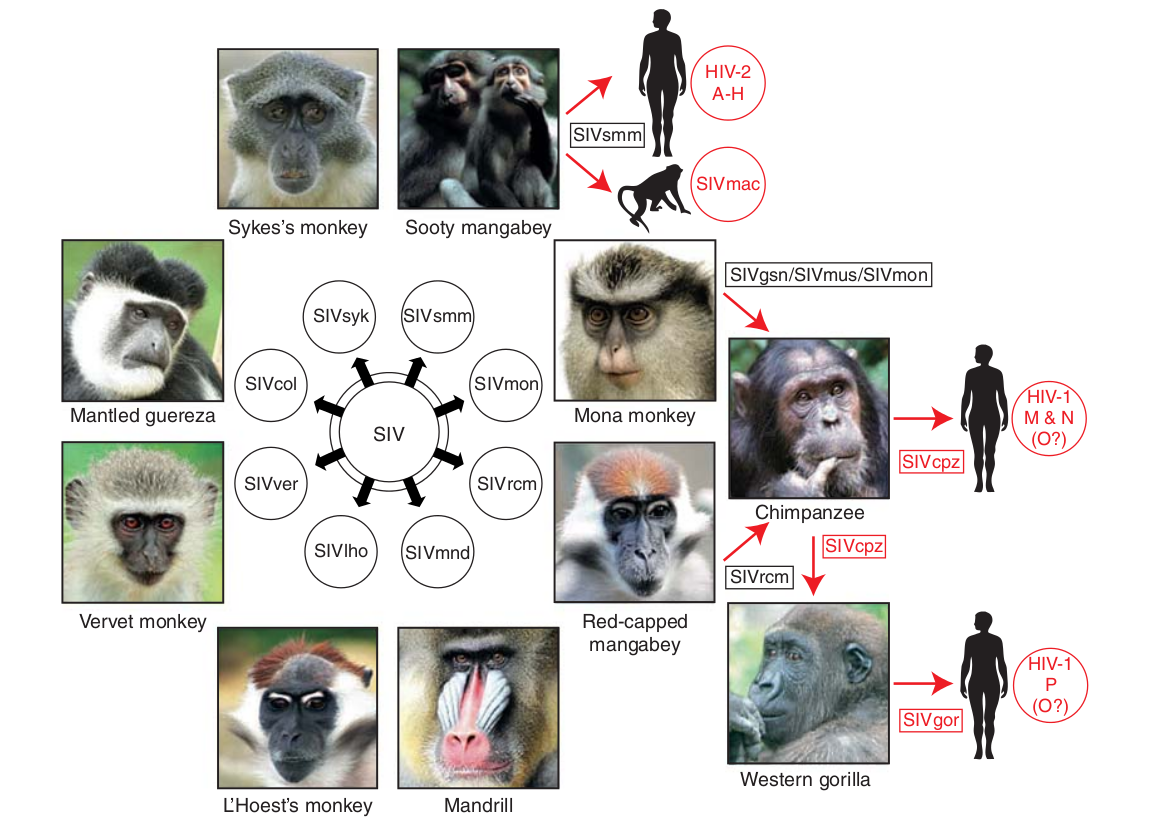
- Chimp → human transmission ~1900 gave rise to HIV-1 group M
- Diversified into subtypes that are ~20% different
- evolves at a rate of about 0.1% per year
HIV infection
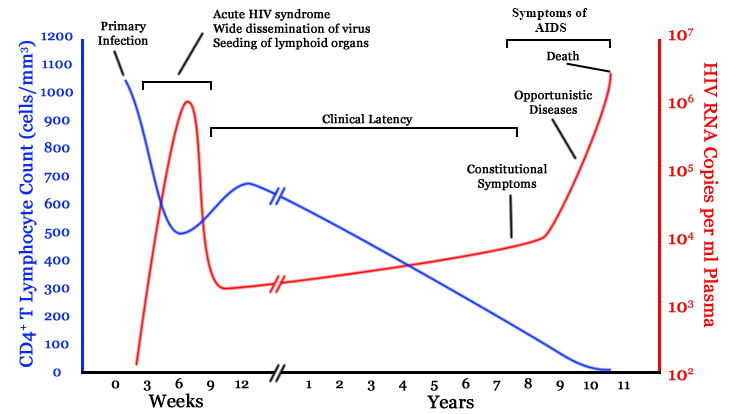 chronic infection:
chronic infection:
- $10^8$ cells are infected every day
- the virus repeatedly escapes immune recognition
- integrates into T-cell as
latent provirus
HIV-1 sequencing before and after therapy
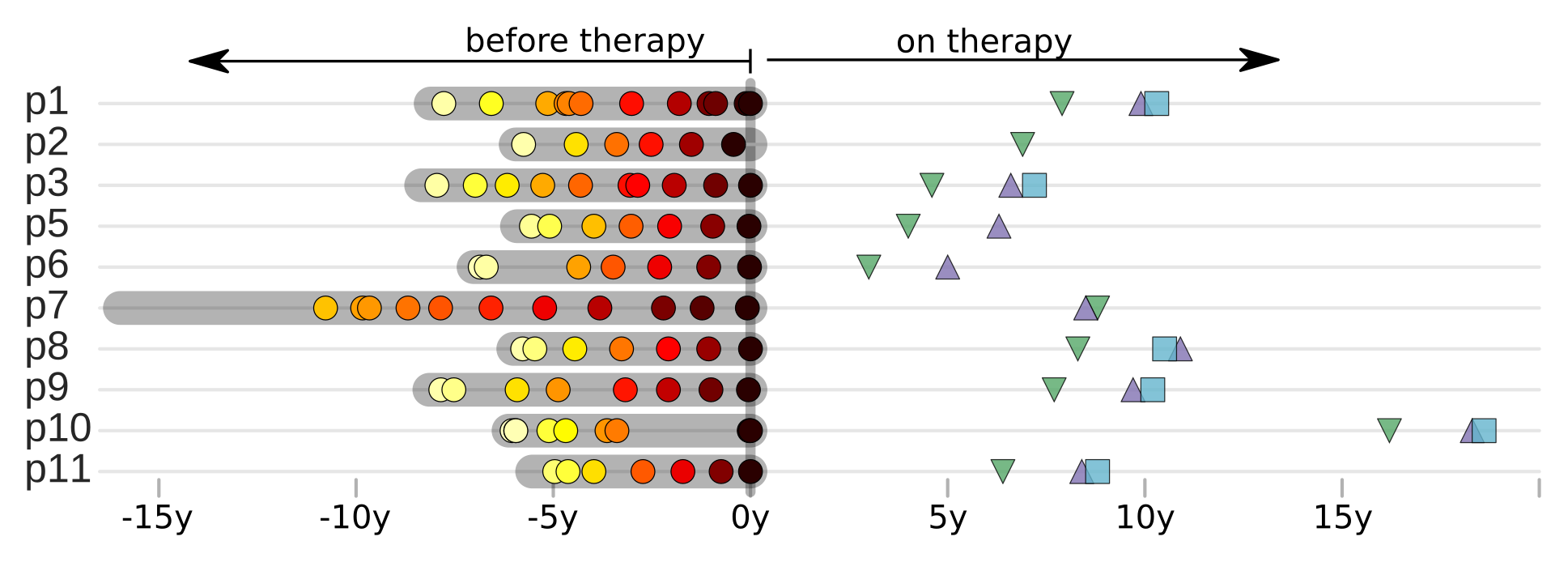 Zanini et al, eLife, 2015;
Brodin et al, eLife, 2016
Zanini et al, eLife, 2015;
Brodin et al, eLife, 2016
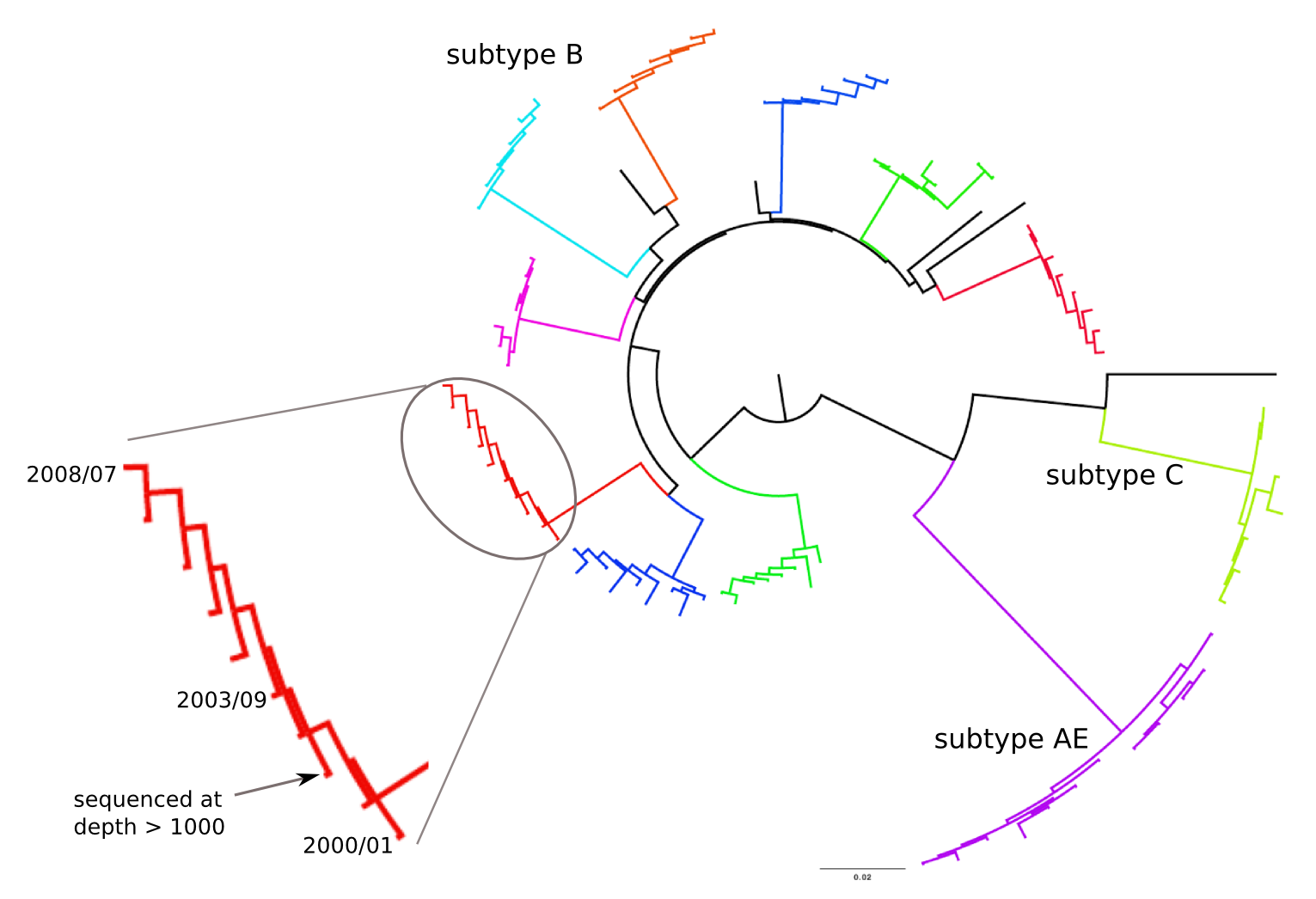
Population sequencing to track all mutations above 1%
- diverge at 0.1-1% per year
- almost full genomes coverage in 10 patients
- full data set at hiv.tuebingen.mpg.de
Diversity and mutation rates
- envelope changes fastest, enzymes lowest
- identical rate of synonymous evolution
- diversity saturates where evolution is fast
- synonymous mutations stay at low frequency
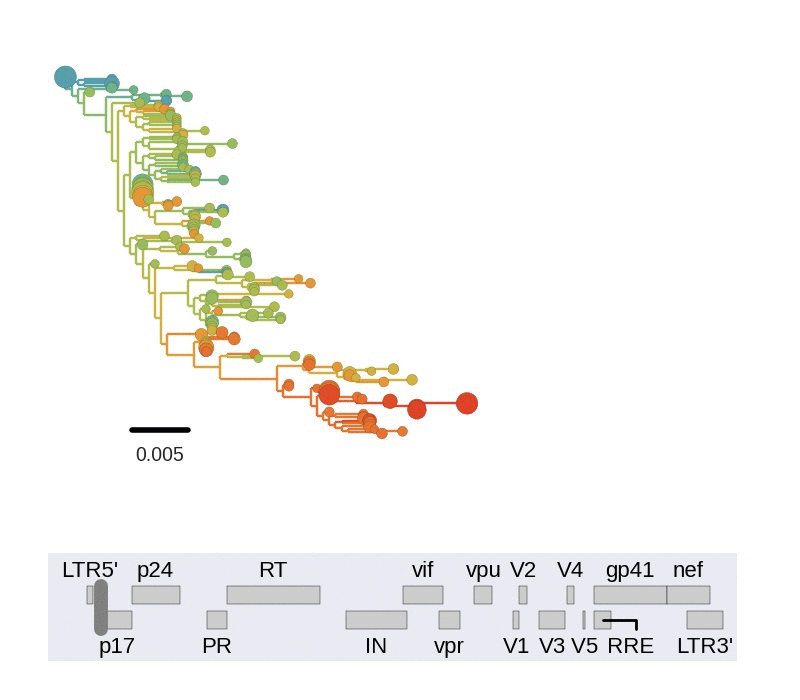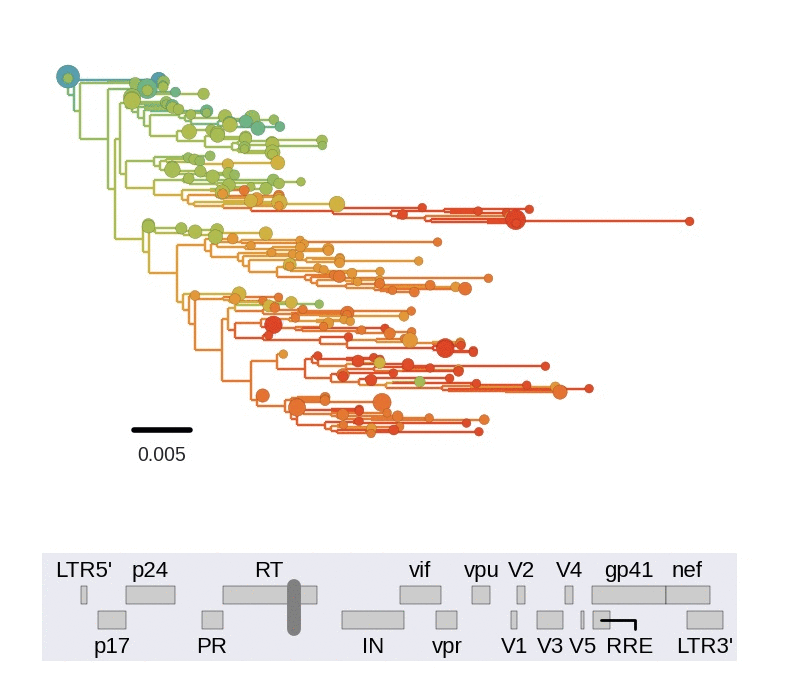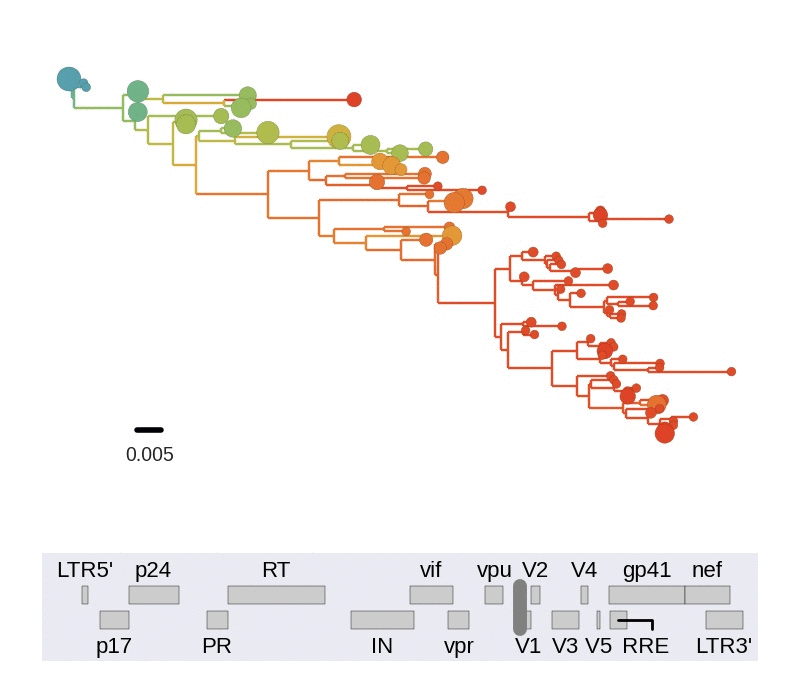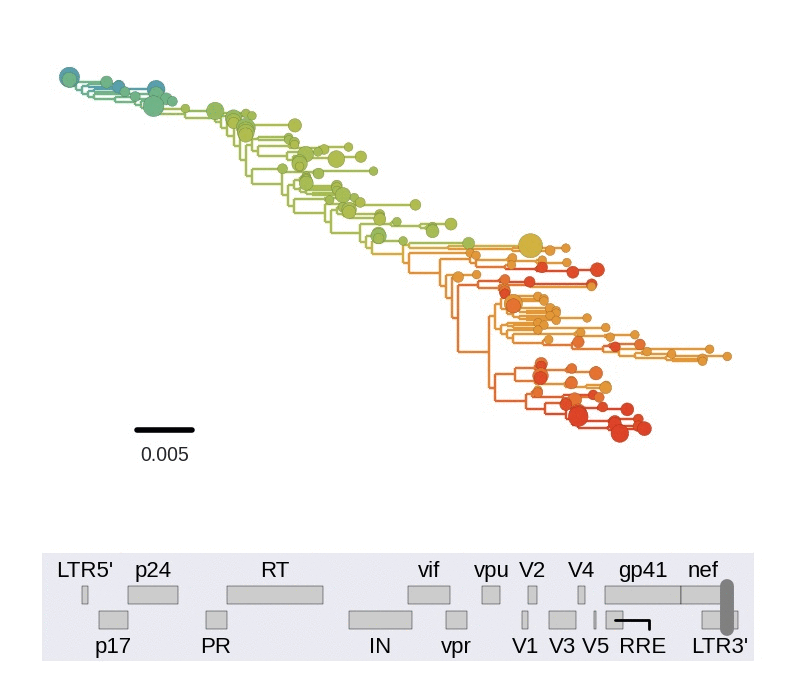
Frequent version of previously beneficial mutations
- HIV escapes immune systems
- most mutations are costly
- humans selects for different mutations
- compensation or reversion?
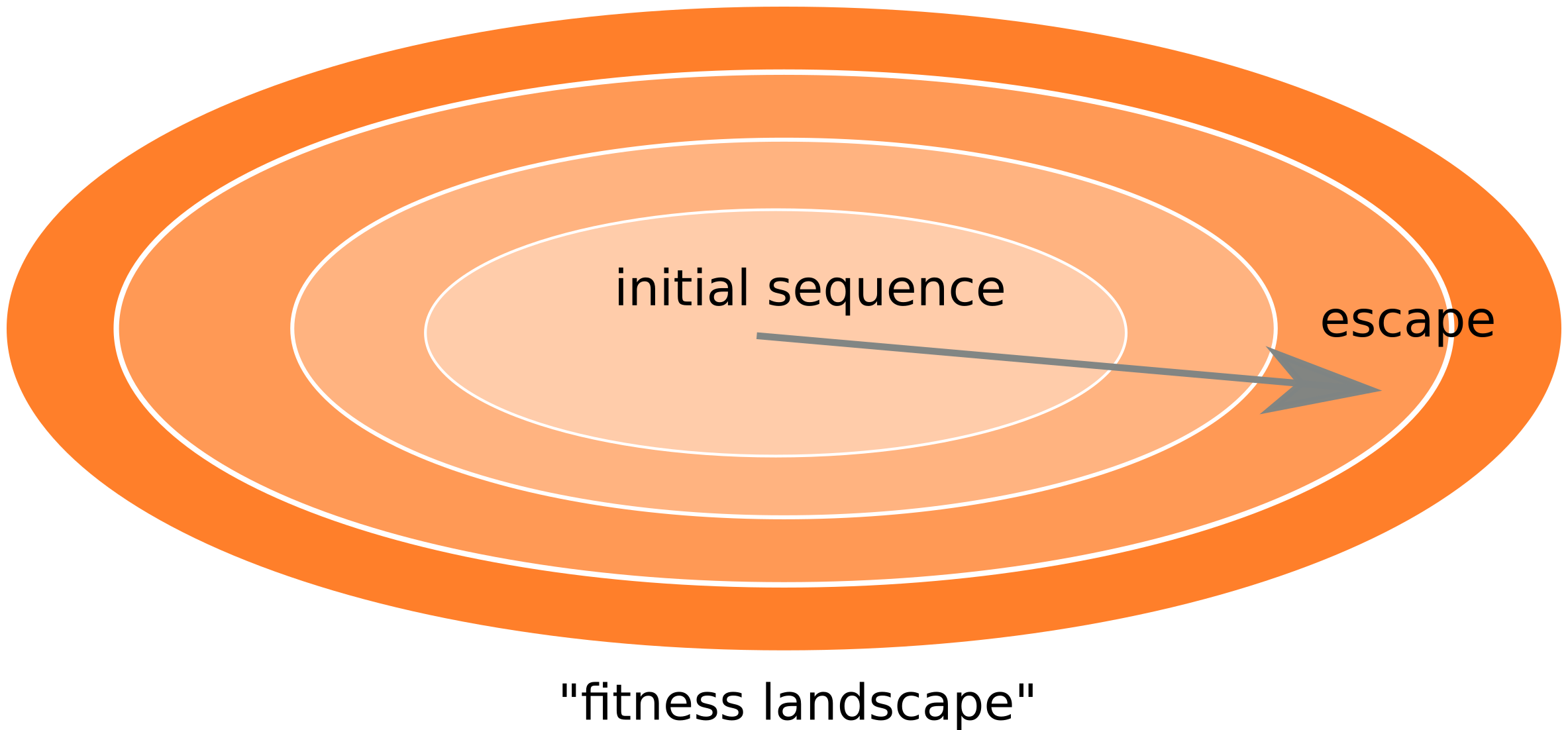
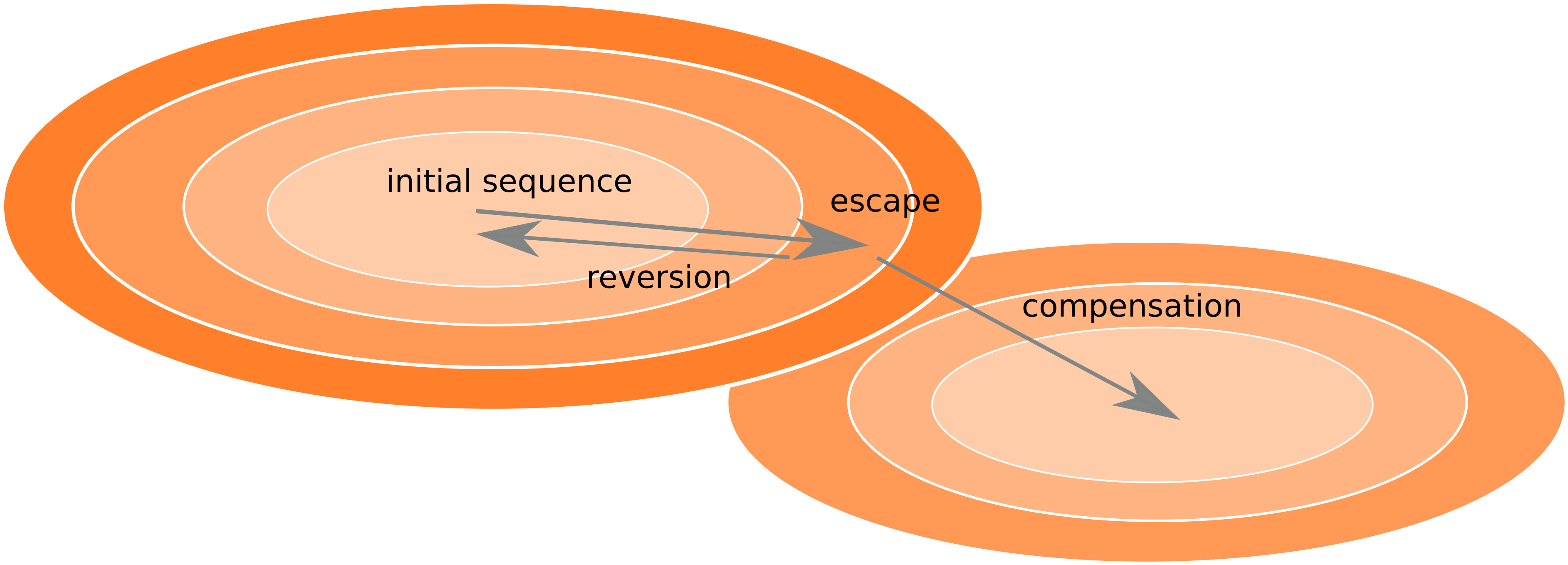
Fitness landscape of HIV-1
Zanini et al, Virus Evolution, 2017Does HIV evolve during therapy?
 Brodin et al, eLife, 2016
Brodin et al, eLife, 2016
No evidence of ongoing replication
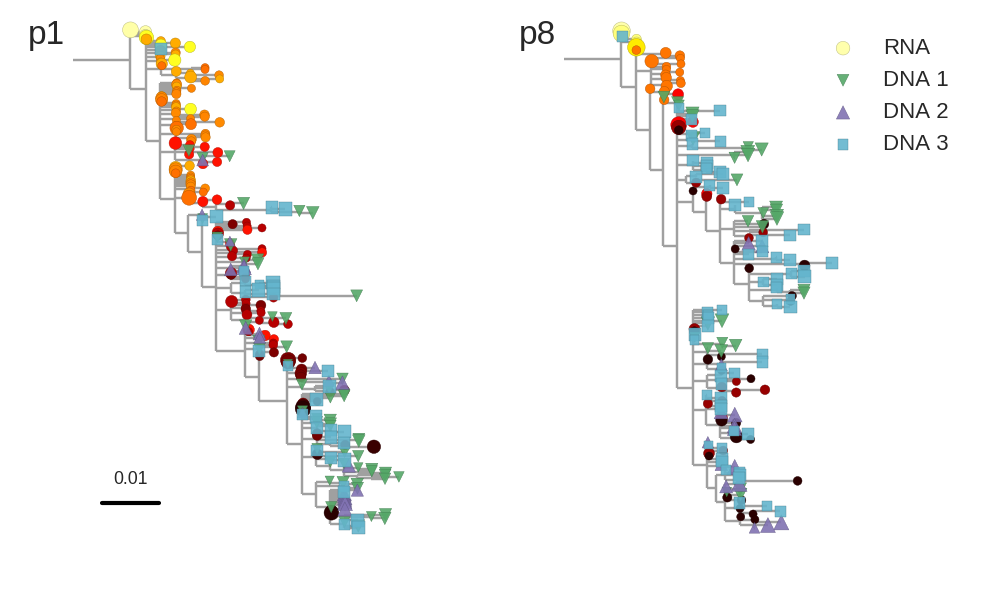
- HIV-1 RNA from plasma before treatment started
- HIV-1 DNA gag-p17 from PBMCs after many years of treatment
No evidence of ongoing replication
T-cell turnover is fast in untreated infection
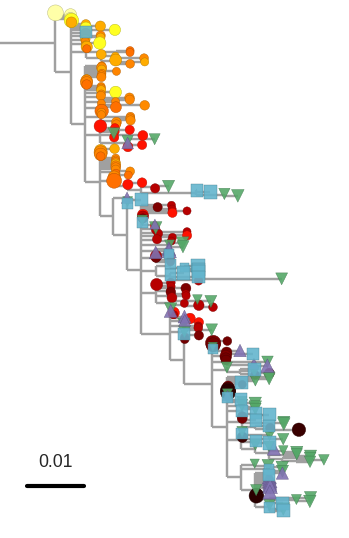
- latent HIV → barcode of a T-cell lineage
- all latent integrated virus derives from late infection
- untreated: T-cell lineages are short lived
- on therapy: T-cell clones live decades
Sharing of viral NGS data is challenging!
- Short reads need to be filtered, mapped, assembled, ...
- Metadata is critical (template input, CD4 counts, etc)
- Processed data much more useful than raw reads
hiv.tuebingen.mpg.de
Horizontal transfer, pangenomes, and bacterial diversity
- bacterial phylogenetics is more complicated!
- much bigger data sets
- horizontal transfer
- messy assemblies
HIV acknowledgments
- Fabio Zanini
- Jan Albert
- Johanna Brodin
- Christa Lanz
- Göran Bratt
- Lina Thebo
- Vadim Puller


Influenza and Theory acknowledgments
- Boris Shraiman
- Colin Russell
- Trevor Bedford


nextstrain.org team
- Colin Megill
- Trevor Bedford
- James Hadfield
- Sidney Bell
the group
