Virus evolution and the predictability of next year's flu
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/201703_Paris.html
Evolution of HIV
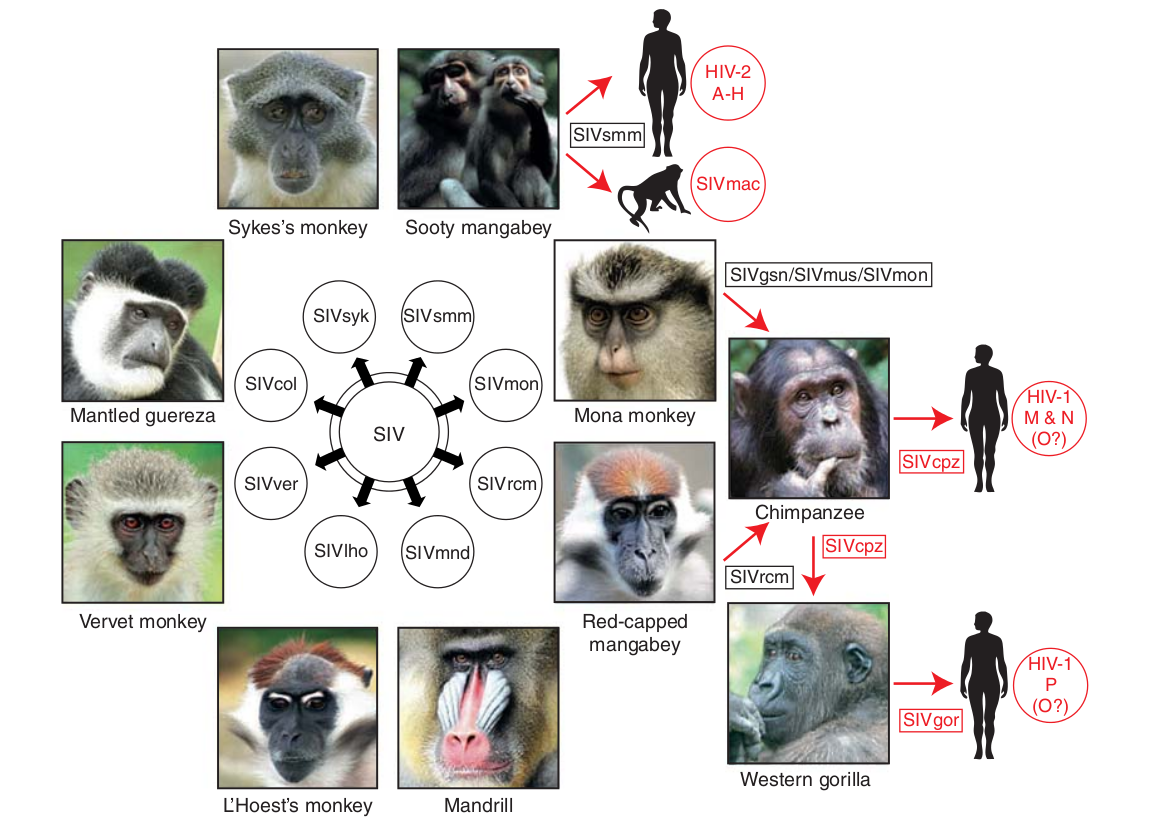
- Chimp → human transmission ~1900 gave rise to HIV-1 group M
- Diversified into subtypes that are ~20% different
- evolves at a rate of about 0.1% per year
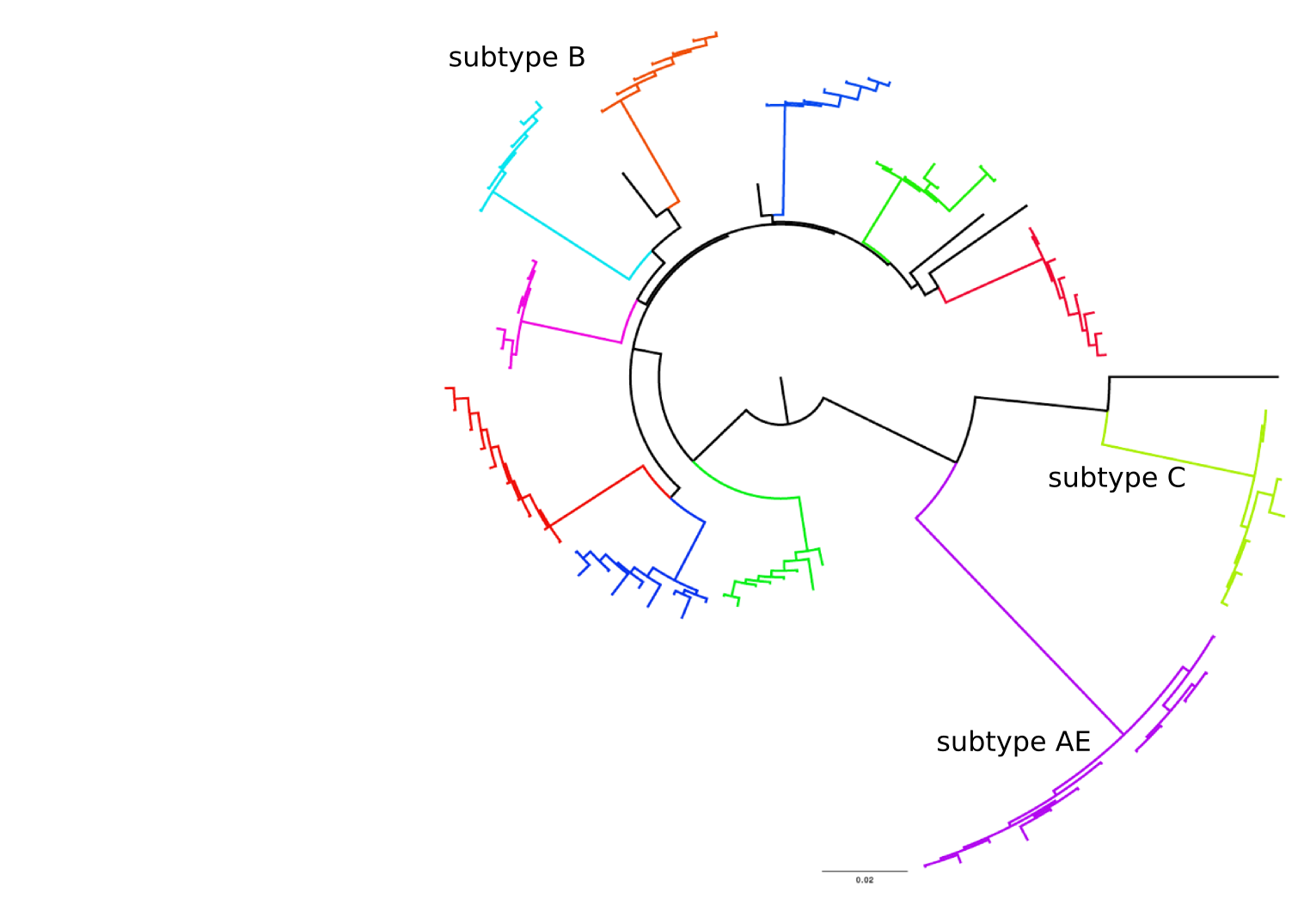
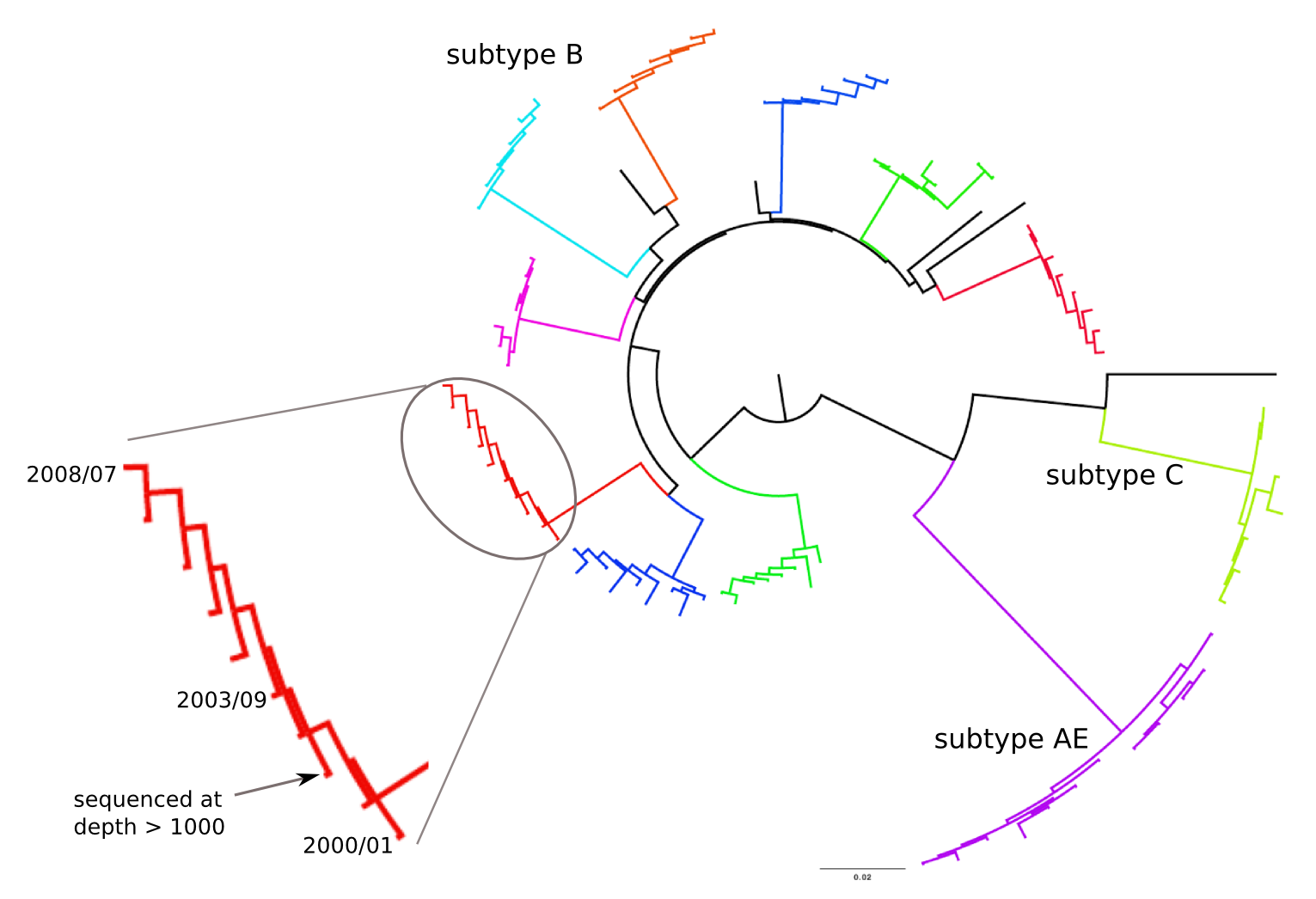
Population sequencing to track all mutations above 1%
- diverge at 0.1-1% per year
- almost full genomes coverage in 10 patients
- full data set at hiv.tuebingen.mpg.de
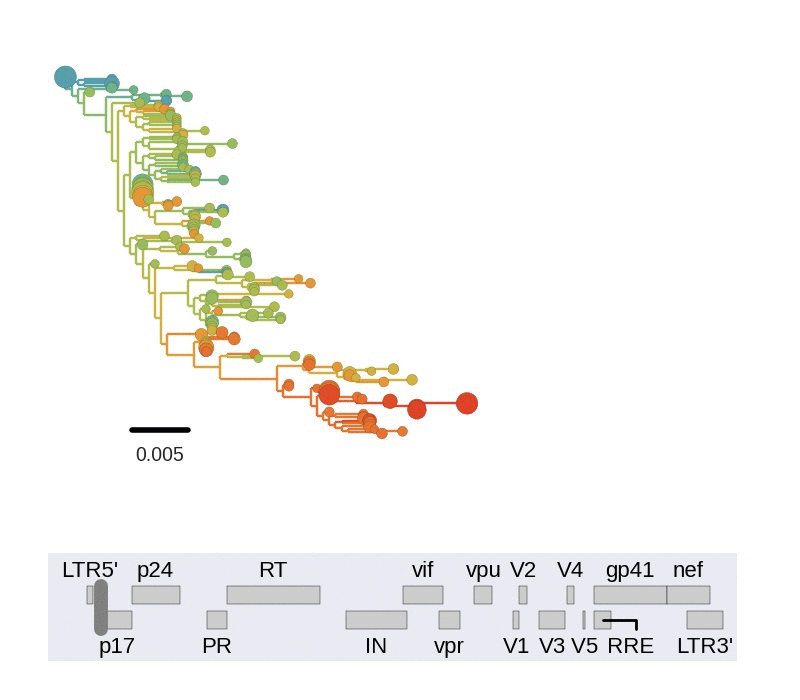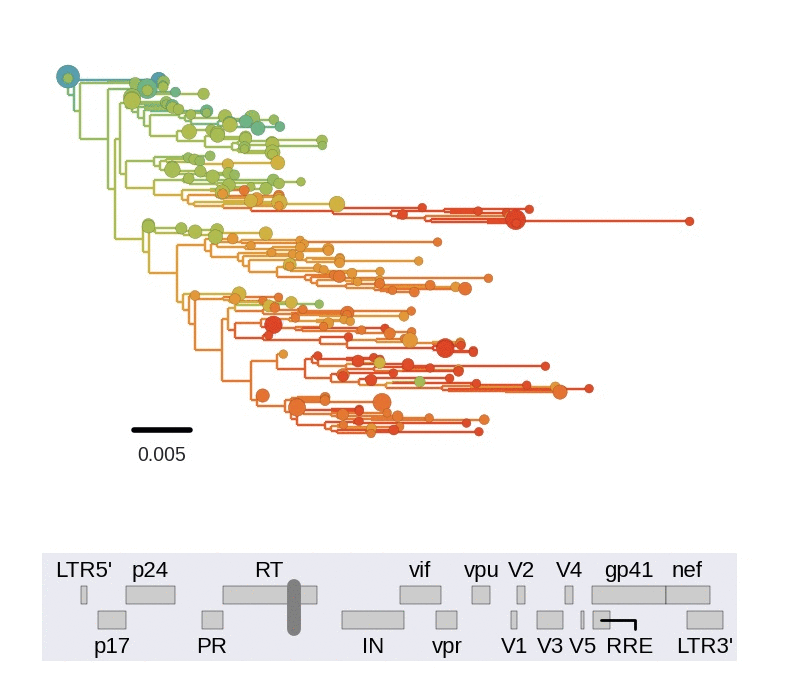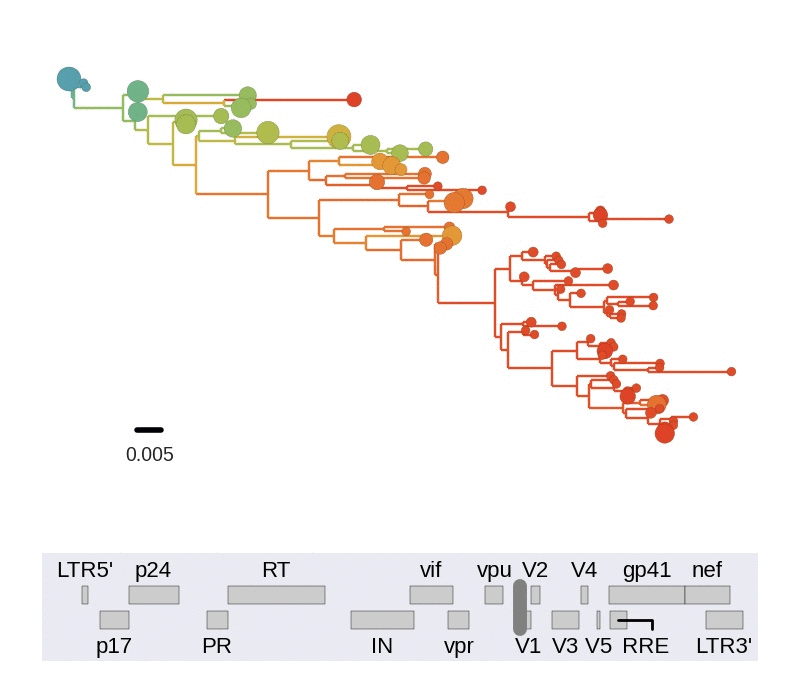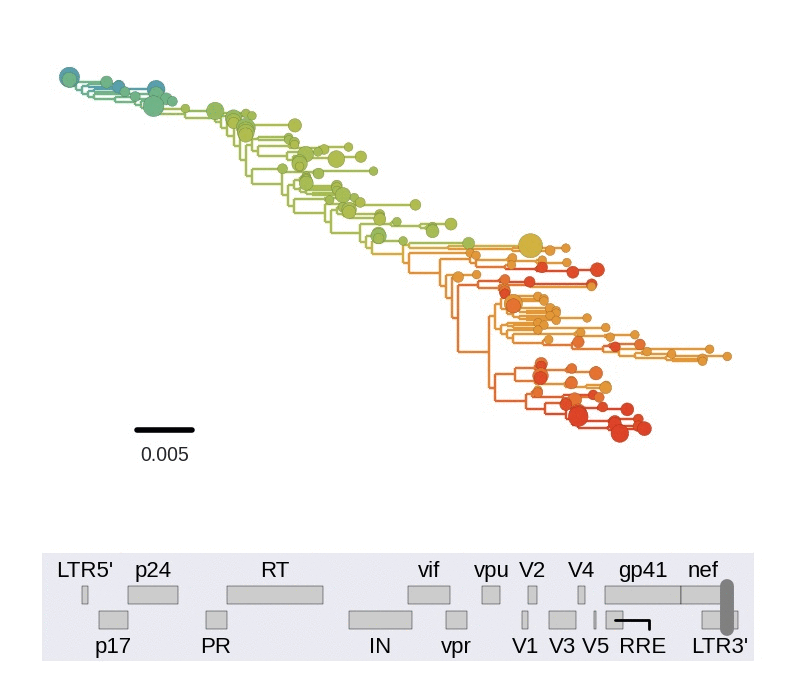
Diversity and hitchhiking
- envelope changes fastest, enzymes lowest
- identical rate of synonymous evolution
- diversity saturates where evolution is fast
- synonymous mutations stay at low frequency
Frequent version of previously beneficial mutations
- HIV escapes immune systems
- most mutations are costly
- humans selects for different mutations
- compensation or reversion?
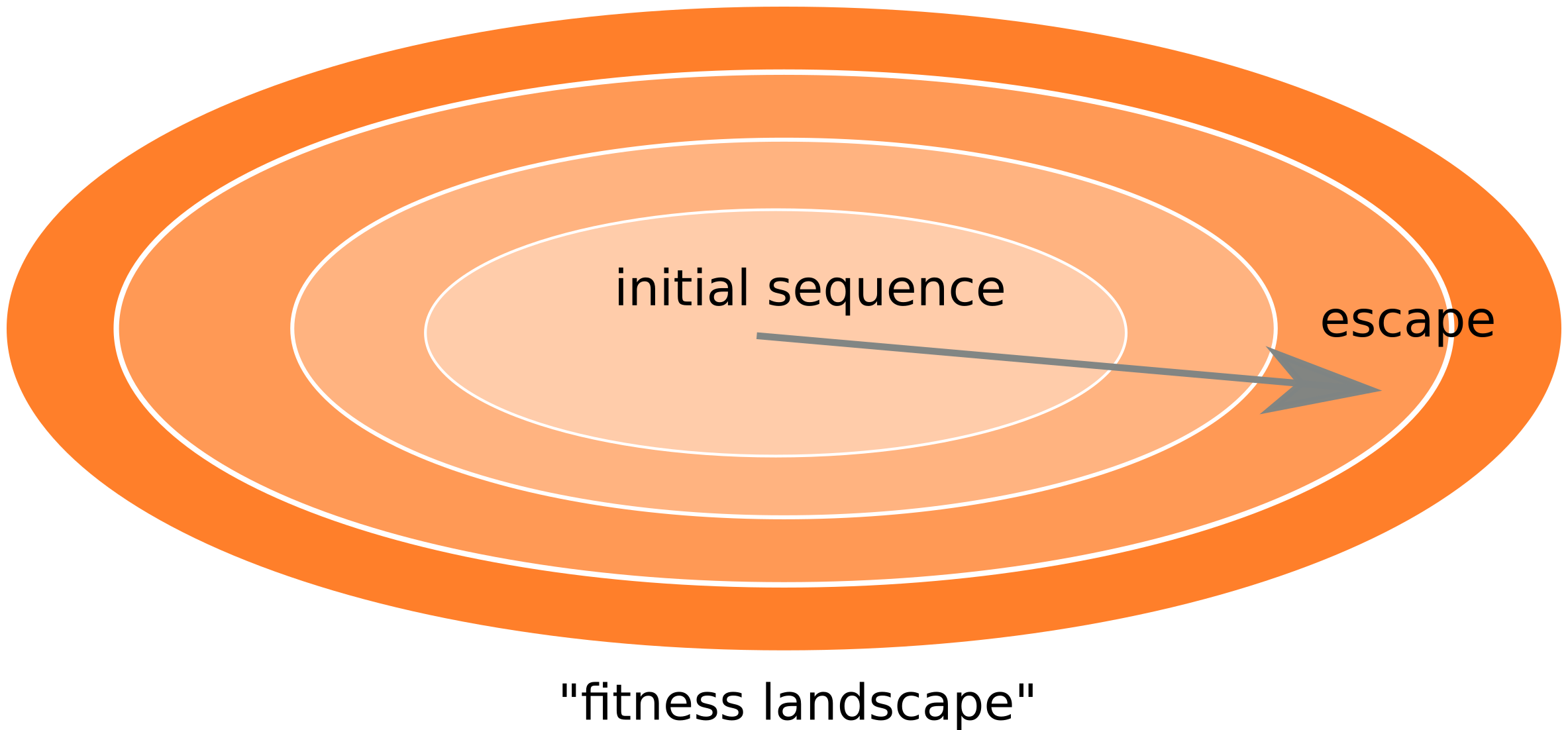
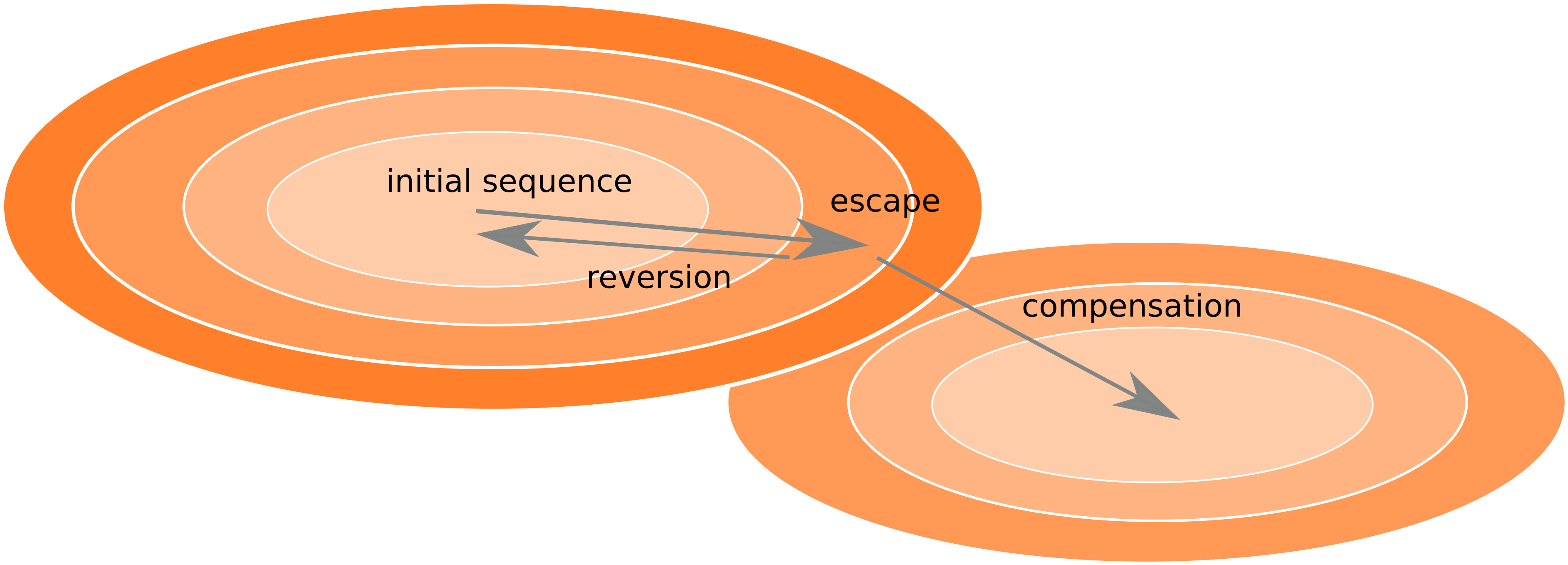
Inference of fitness costs
- mutation away from preferred state with rate $\mu$
- selection against non-preferred state with strength $s$
- variant frequency dynamics: $\frac{d x}{dt} = \mu -s x $
- equilibrium frequency: $\bar{x} = \mu/s $
- fitness cost: $s = \mu/\bar{x}$
Fitness landscape of HIV-1
Zanini et al, Virus Evolution, 2017Selection on RNA structures and regulatory sites
Zanini et al, Virus Evolution, 2017The distribution of fitness costs
Zanini et al, Virus Evolution, 2017Fitness - diversity correlation
Zanini et al, Virus Evolution, 2017Population genetics models
evolutionary processes ↔ trees ↔ genetic diversity
Neutral models and beyond
Neutral models
- all individuals are identical → same offspring distribution
- Kingman coalesence and diffusion theory are dual descriptions
- everything is easy to calculate
- perturbations like background selection can be included
What if selection is everywhere?
Clonal interference and traveling waves
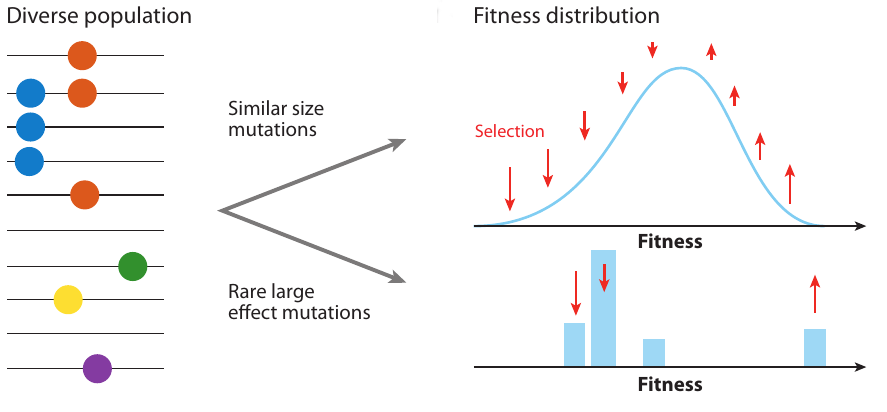
- extensive work on speed of adaptation, but this speed is not observable
- genetic diversity is what we observe
- depends on the properties of trees
What if selection is everywhere?
Kingman coalescent
strong selection
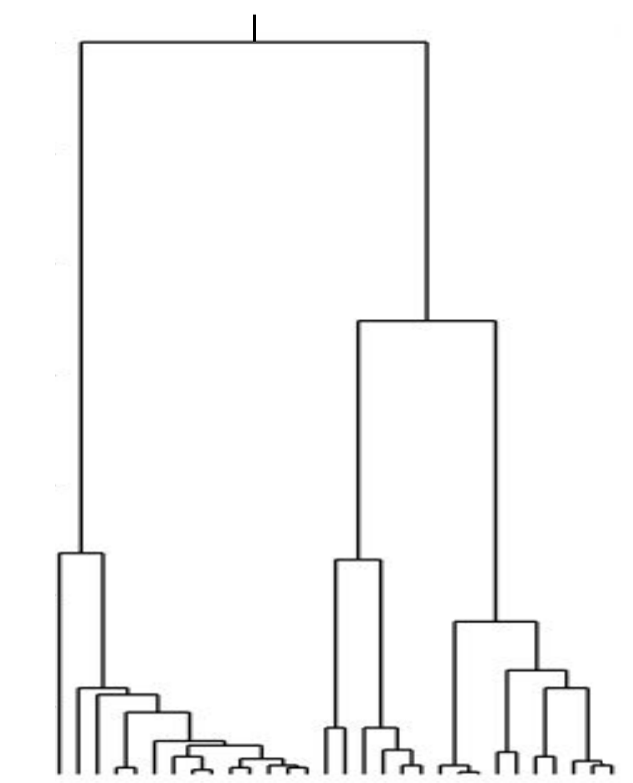

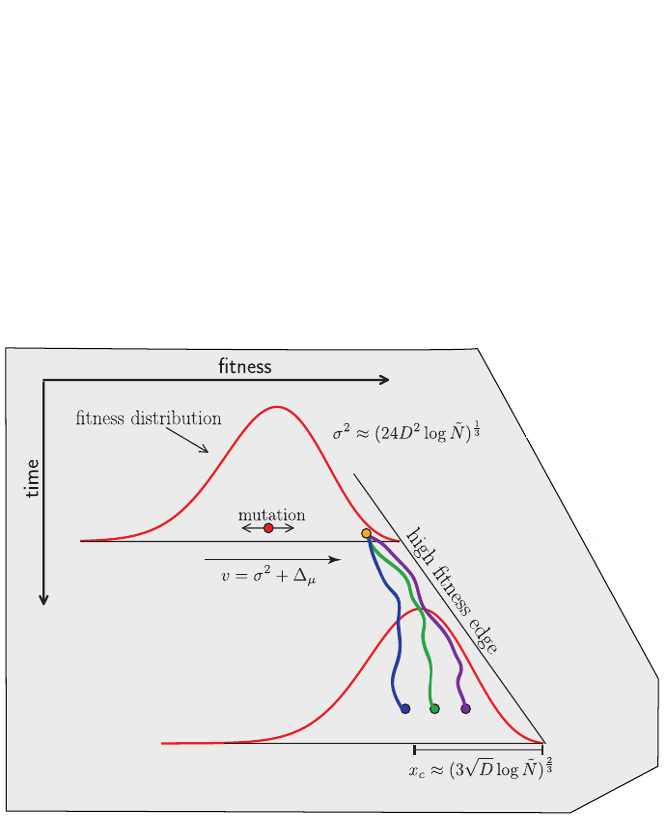
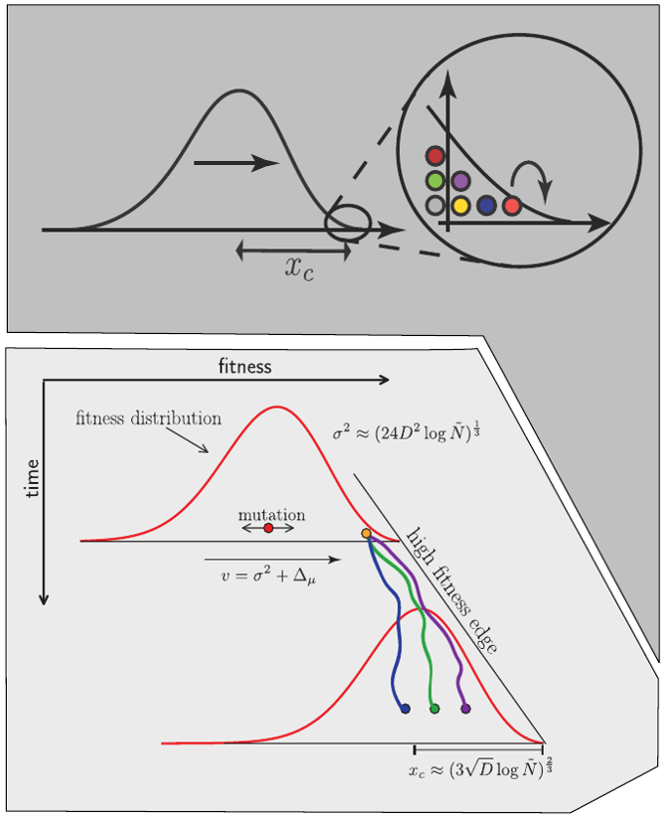
Bolthausen-Sznitman Coalescent
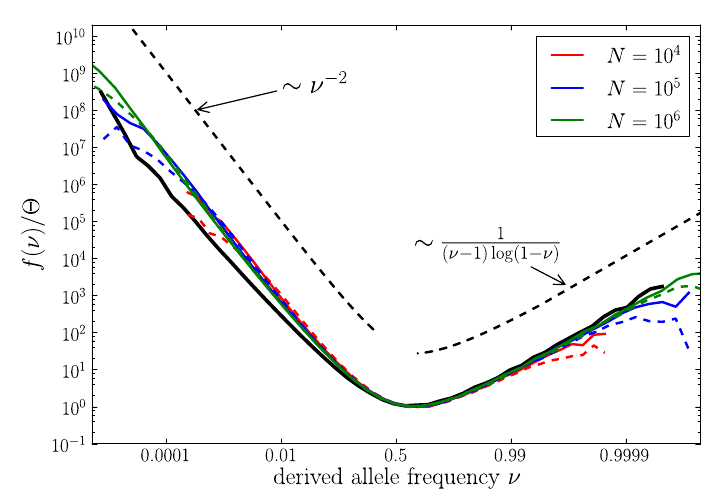
U-shaped polarized site frequency spectra

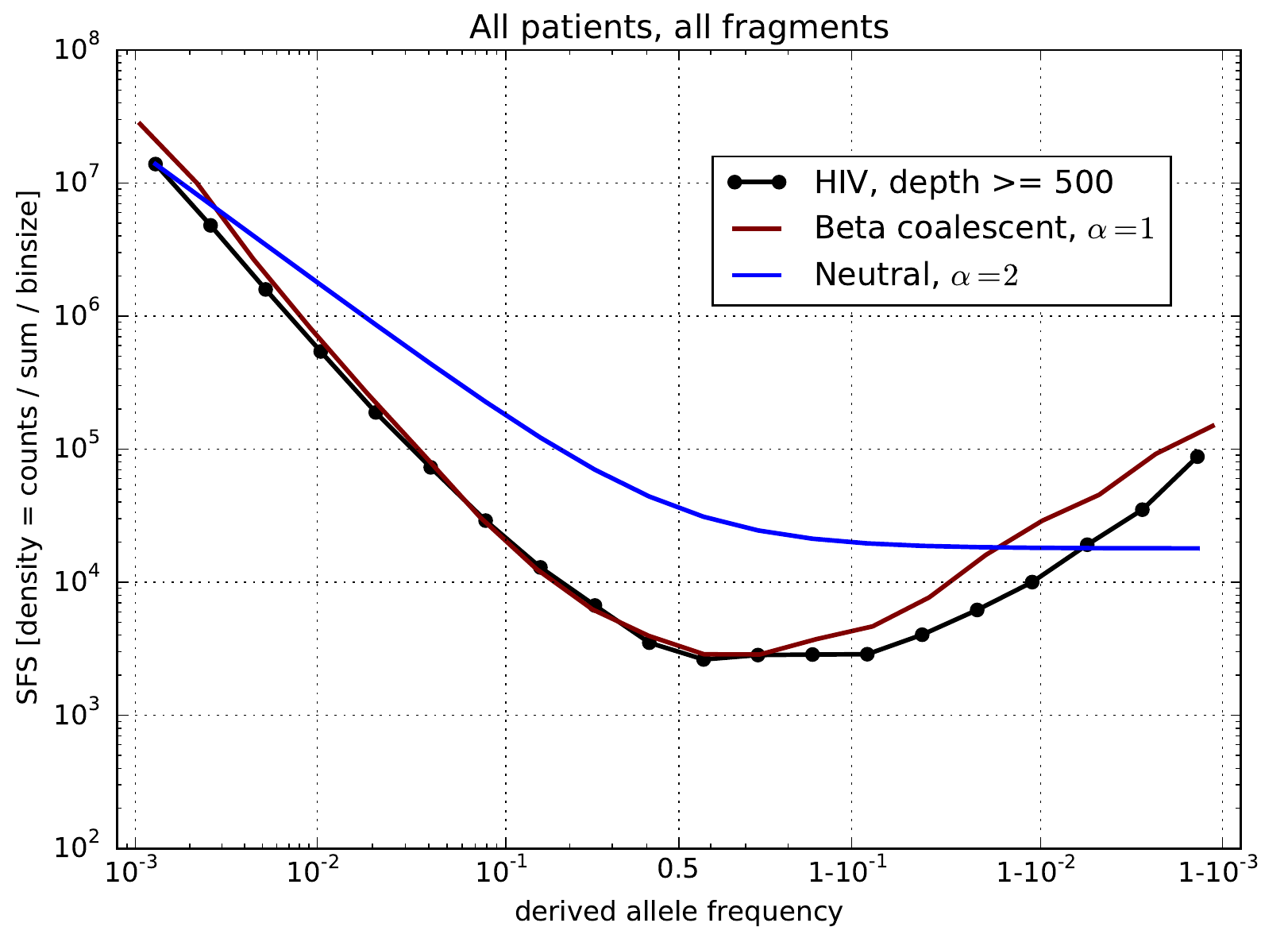 Zanini et al, eLife, 2015
Zanini et al, eLife, 2015
Bursts in a tree ↔ high fitness genotypes
Can we read fitness of a tree?



- Influenza virus evolves to avoid human immunity
- Vaccines need frequent updates

Predicting evolution

Given the branching pattern,
RN, Russell, Shraiman, eLife, 2014
- can we predict fitness?
- pick the closest relative of the future?
Fitness inference from trees
$$P(\mathbf{x}|T) = \frac{1}{Z(T)} p_0(x_0) \prod_{i=0}^{n_{int}} g(x_{i_1}, t_{i_1}| x_i, t_i)g(x_{i_2}, t_{i_2}| x_i, t_i)$$
RN, Russell, Shraiman, eLife, 2014
Validate on simulation data
- simulate evolution
- sample sequences
- reconstruct trees
- infer fitness
- predict ancestor of future
- compare to truth

Validation on simulated data
Validation on simulated data
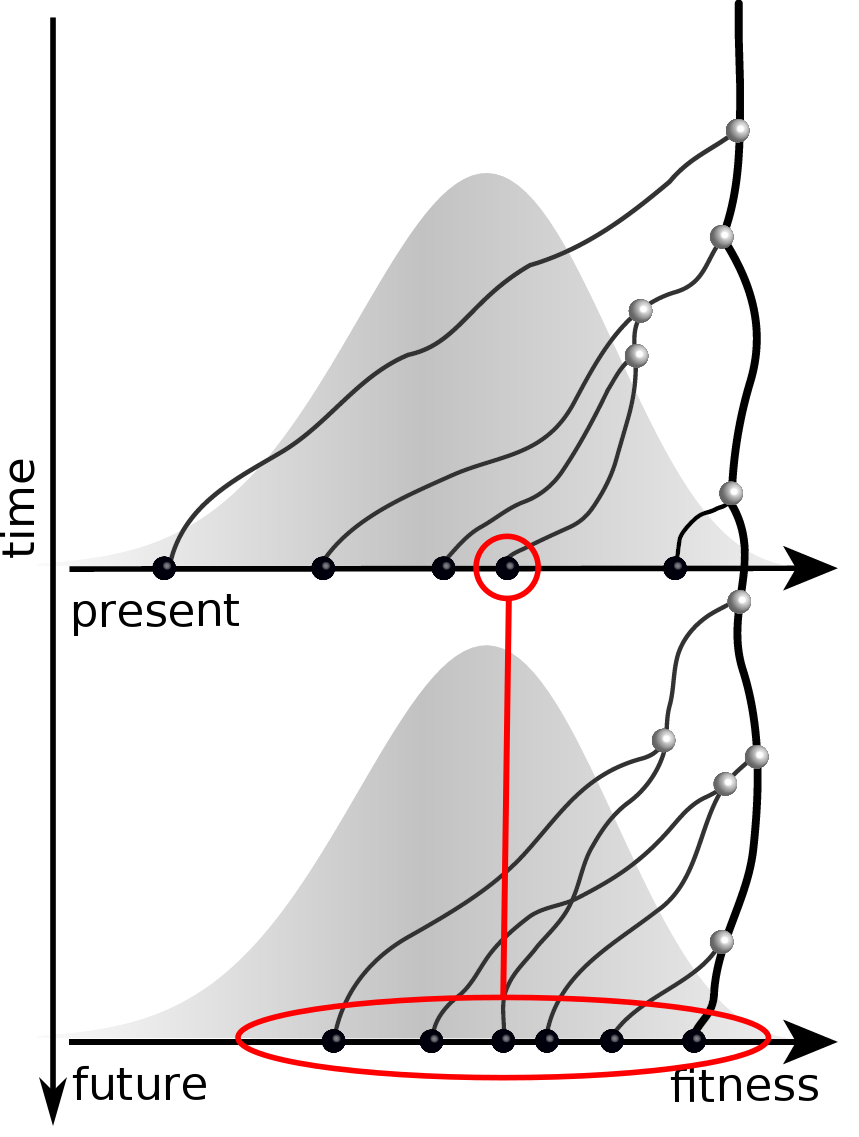
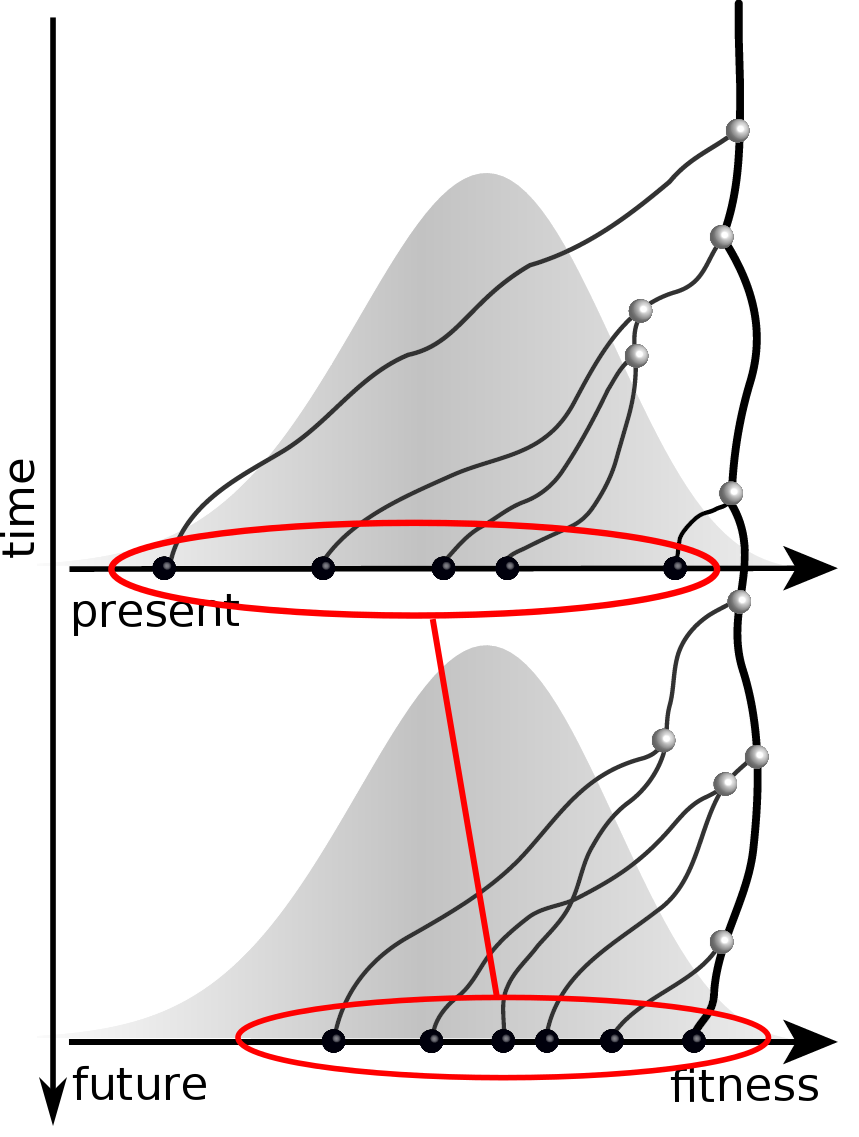
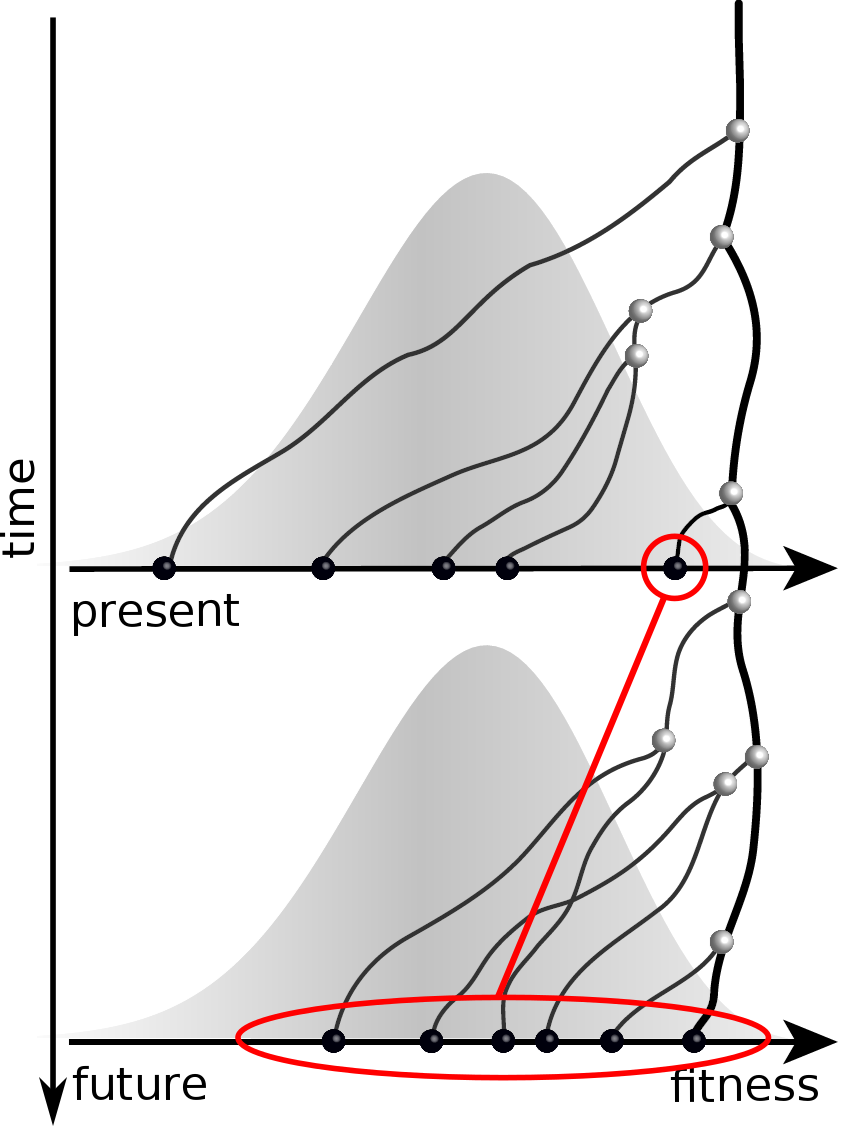
Prediction of the dominating H3N2 influenza strain
RN, Russell, Shraiman, eLife, 2014HIV acknowledgments
- Fabio Zanini
- Jan Albert
- Johanna Brodin
- Christa Lanz
- Göran Bratt
- Lina Thebo
- Vadim Puller


Influenza and Theory acknowledgments




- Boris Shraiman
- Colin Russell
- Trevor Bedford
- Oskar Hallatschek


We recently moved to the Biozentrum Basel
