Estimating the in-vivo fitness landscape of HIV-1
Fabio Zanini, Vadim Puller, Jan Albert, and Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/201705_HIV_dyn.html
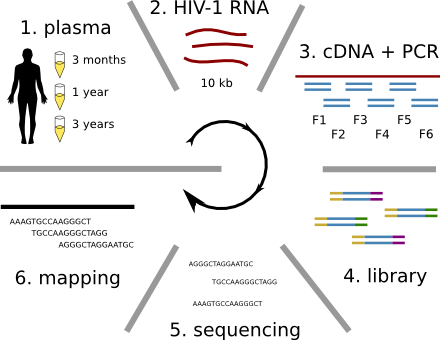
HIV-1 sequencing before and after therapy
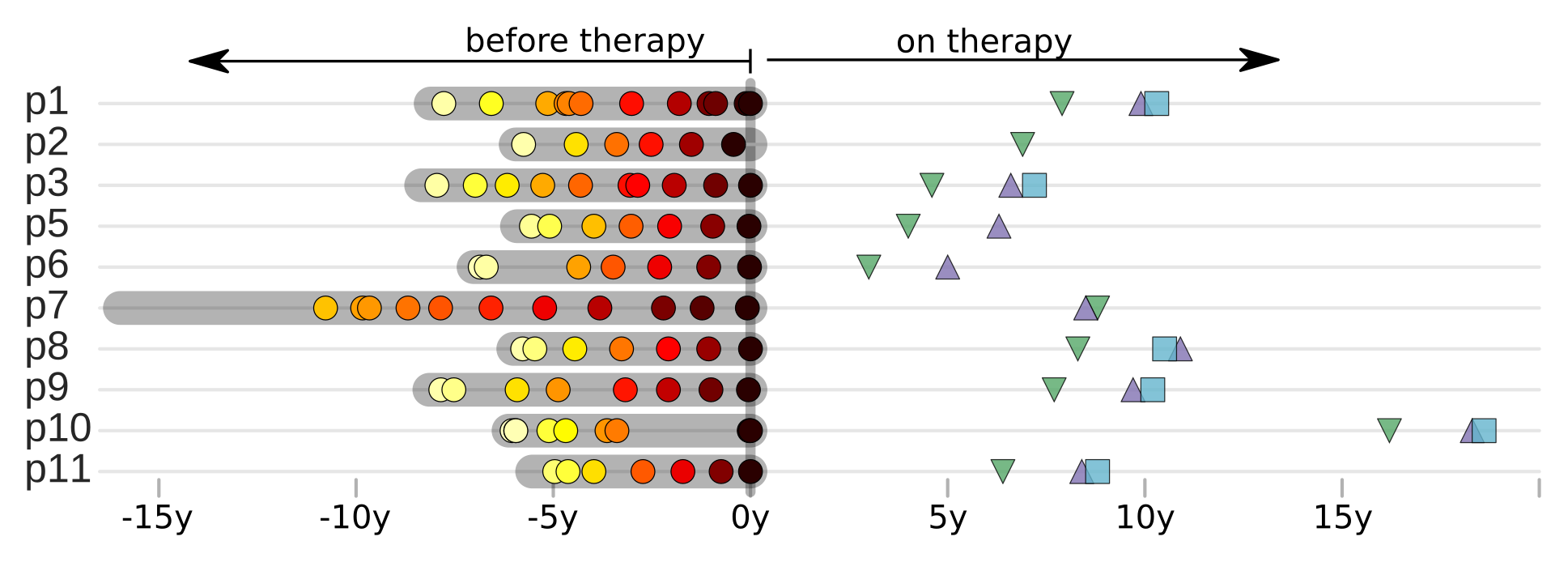 Zanini et al, eLife, 2015;
Brodin et al, eLife, 2016.
Collaboration with the group of Jan Albert
Zanini et al, eLife, 2015;
Brodin et al, eLife, 2016.
Collaboration with the group of Jan Albert
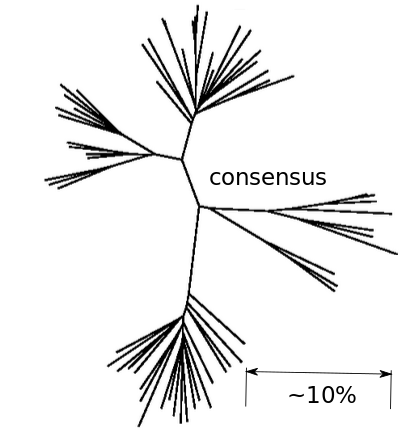
Population sequencing to track all mutations above 1%
- diverge at 0.1-1% per year
- almost full genomes coverage in 10 patients
- full data set at hiv.biozentrum.unibas.ch
Dynamics of single nucleotide variants
- the average frequency of neutral variants grows as $\mu t$
- individual trajectories of SNVs are very noisy
- many sites $\Rightarrow$ average mutation rates
- ASSUMPTION: synonymous and globally variable $\Rightarrow$ approximately neutral
Mutation rates and diversity and neutral sites
Inferred vs measured mutations rates (Abram et al)
Inference of fitness costs
- mutation away from preferred state at rate $\mu$
- selection against non-preferred state with strength $s$
- variant frequency dynamics: $\frac{d x}{dt} = \mu -s x $
- equilibrium frequency: $\bar{x} = \mu/s $
- fitness cost: $s = \mu/\bar{x}$
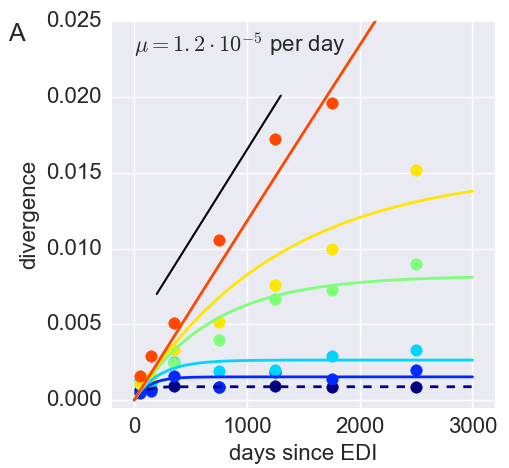
Inference of fitness costs
- Frequencies of costly mutations decorrelate fast $\frac{d x}{dt} = \mu -s x $
- $\Rightarrow$ average many samples to obtain accurate estimates
- Assumption: The global consensus is the preferred state
- Only use sites that initially agree with consensus
- Only use sites that don't chance majority nucleotide
Fitness landscape of HIV-1
Zanini et al, Virus Evolution, 2017Selection on RNA structures and regulatory sites
Zanini et al, Virus Evolution, 2017The distribution of fitness costs
Zanini et al, Virus Evolution, 2017Fitness - diversity correlation
Zanini et al, Virus Evolution, 2017Costly HLA associated positions have high diversity
Zanini et al, Virus Evolution, 2017HIV acknowledgments
- Fabio Zanini
- Jan Albert
- Johanna Brodin
- Vadim Puller


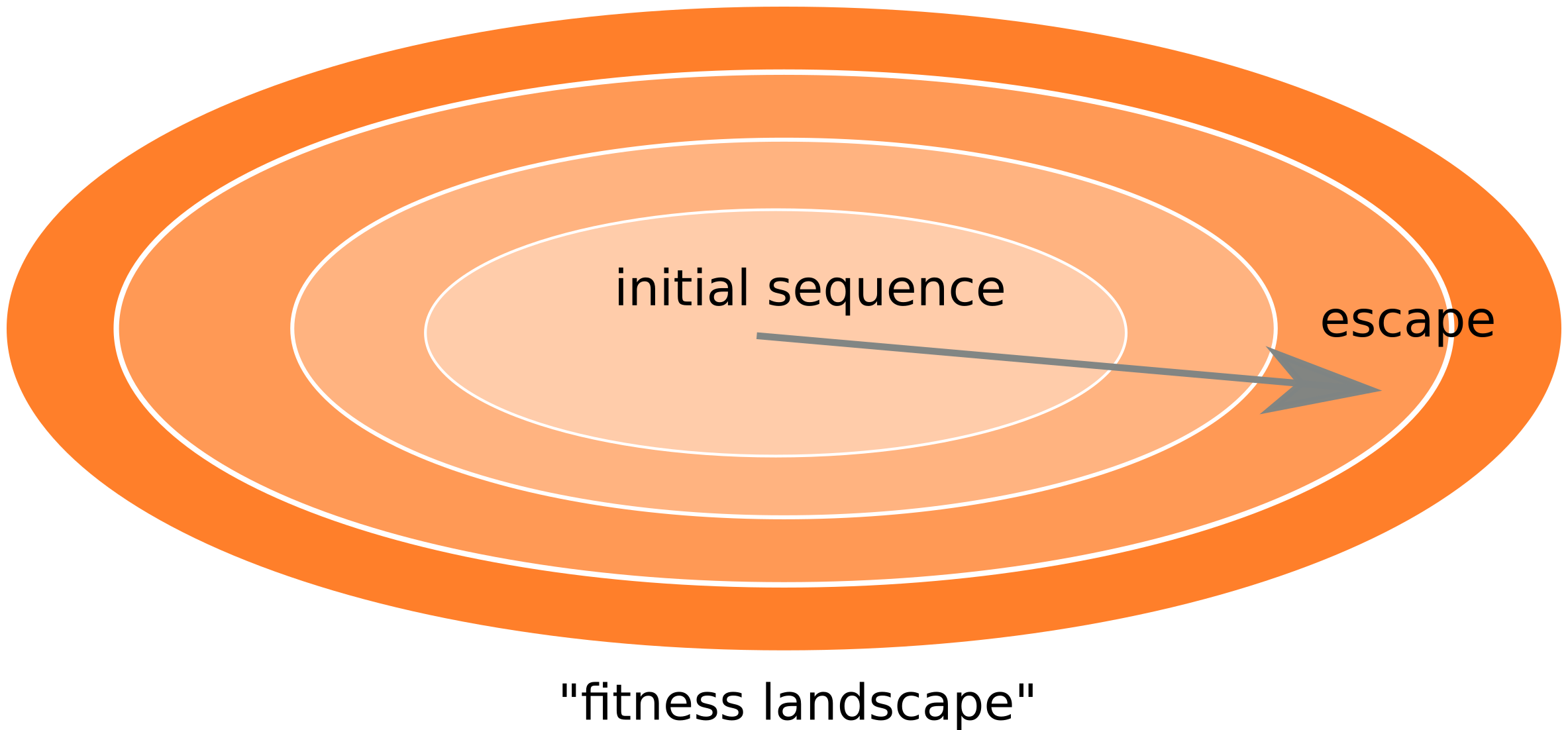
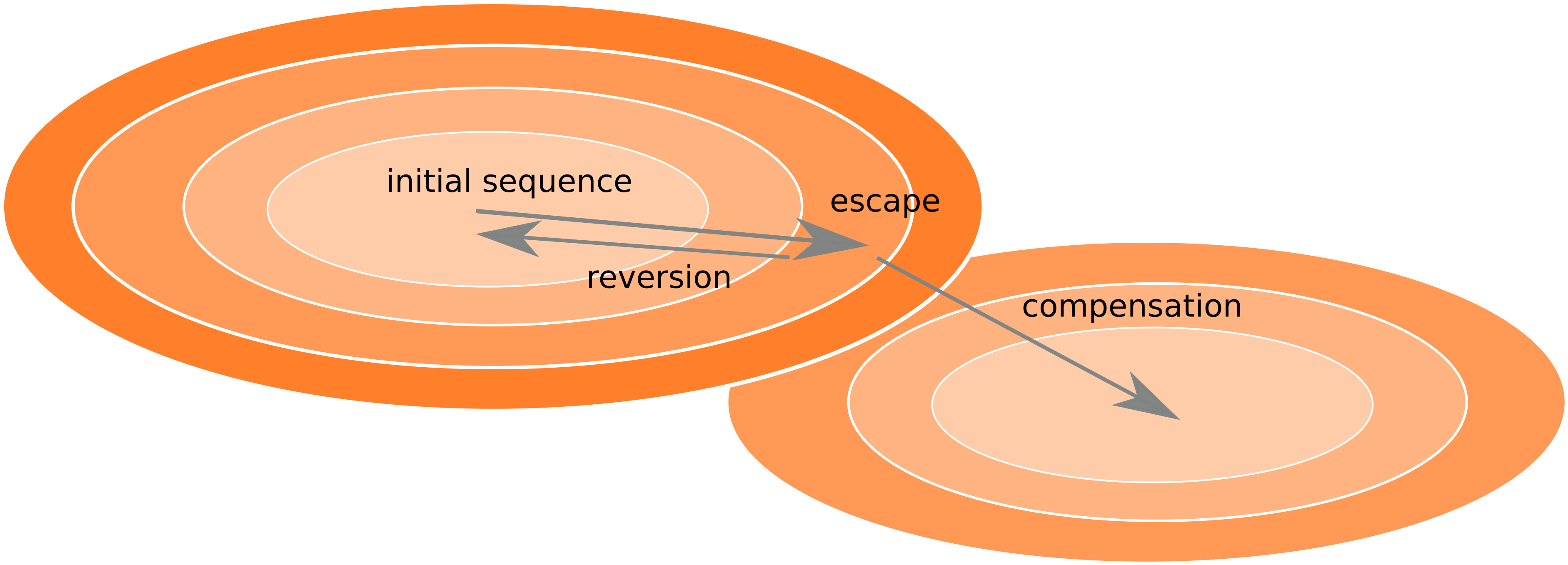
Frequent reversion of previously beneficial mutations
- HIV escapes immune systems
- most mutations are costly
- humans selects for different mutations
- compensation or reversion?
Deep sequencing of plasma RNA