Real-time tracking of seasonal influenza, Ebola, and Zika viruses
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/201706_RKI.html
Evolution of RNA viruses
- Constant struggle to adapt to changing environments and host immunity
- Mutation rates of about 0.00001/site and replication
- Large viral populations explore every single mutation every day
- Most mutations are detrimental, but some persist
- Mutations can act like an approximate clock
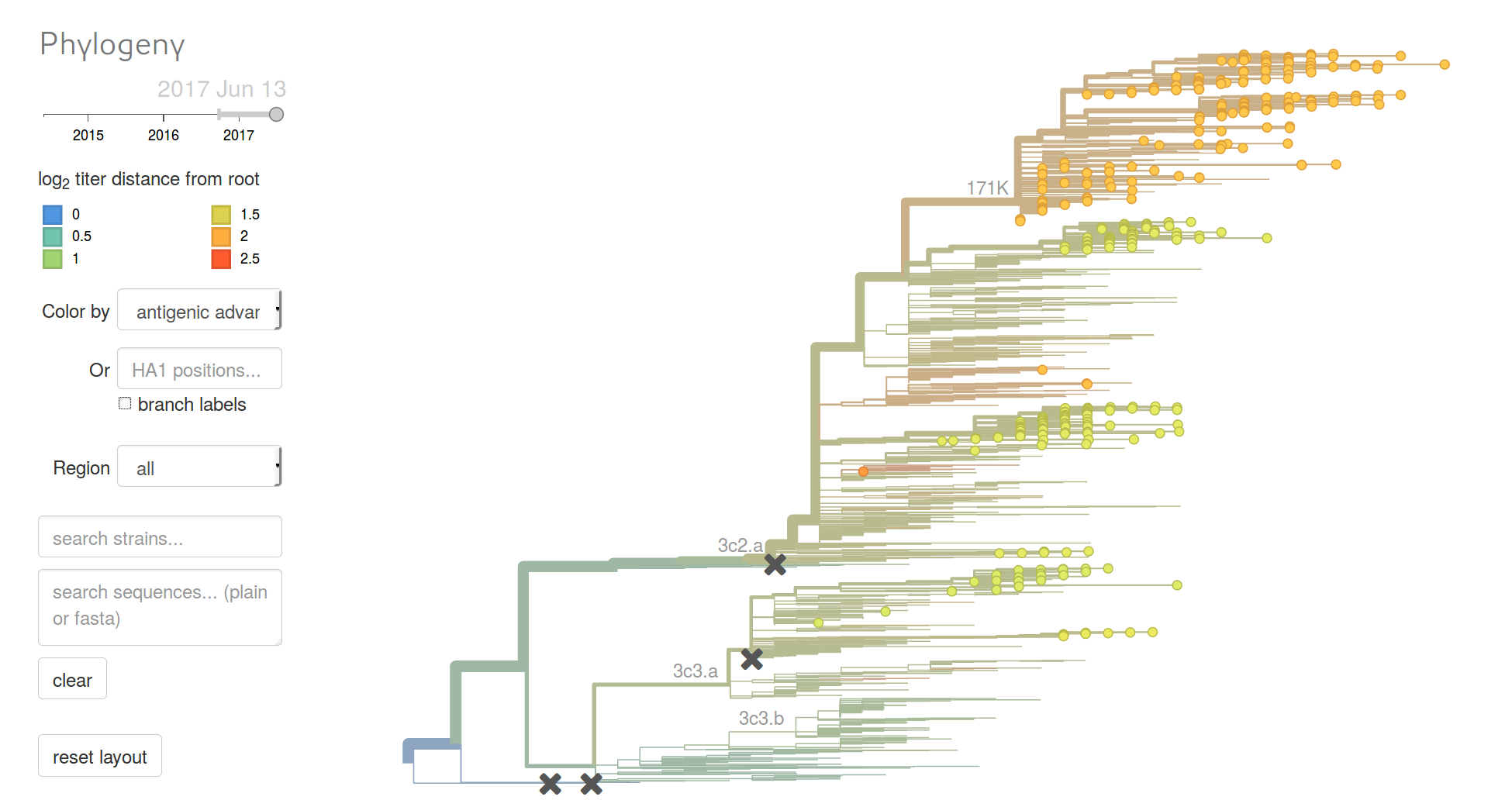
Phylogenetic analysis
Phylogenetic analysis
Can we use sequences to track an ongoing outbreak?
Can we predict future evolution of influenza viruses?
Sequences record the spread of pathogens




The resolution is limited by the number of mutations!
Influenza virus genome - 8 segments
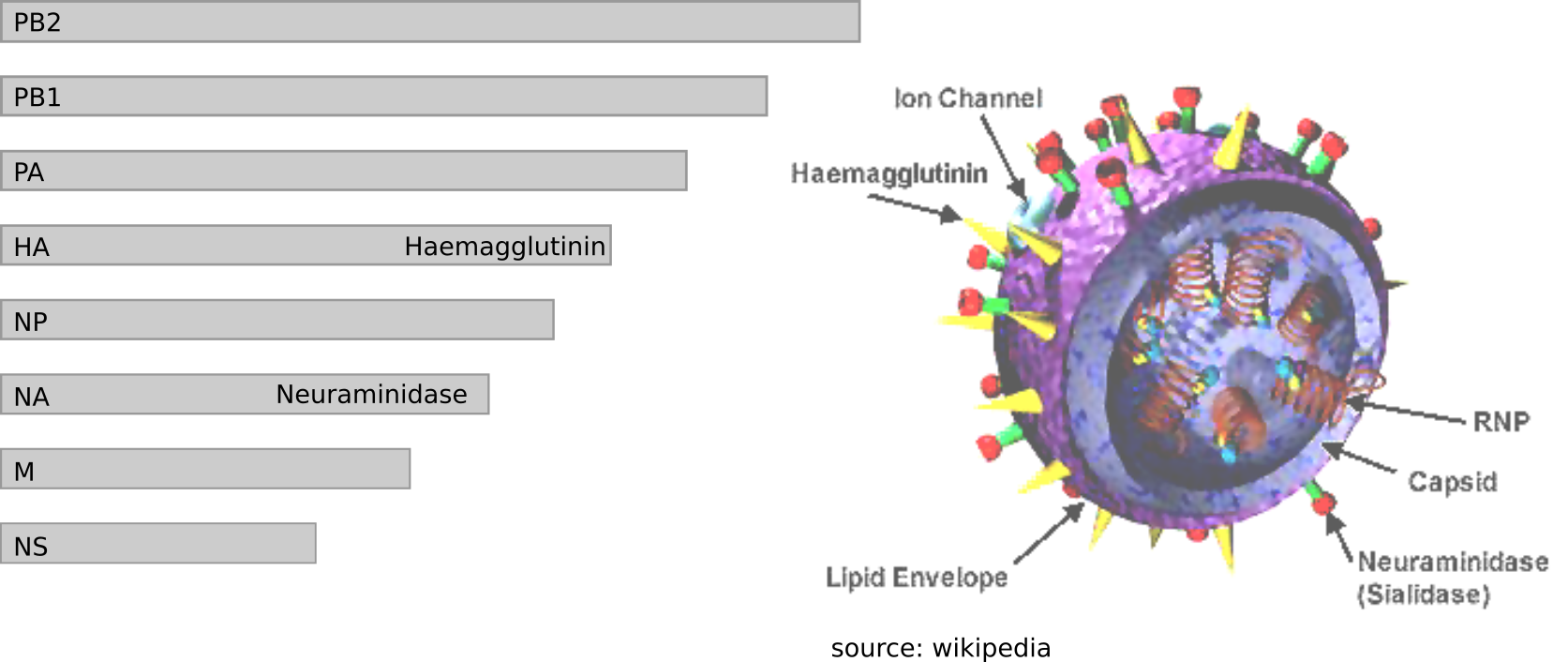
Zika virus genome $\sim 10000$ bases

Ebola virus genome $\sim 20000$ bases

Many RNA viruses pick up one mutation every 2-4 weeks!
Frequent mutations imply...
- most viruses in an outbreak differ from each other
- transmission chains are visible
- transmission can be ruled out!
- geographic spread can be reconstructed
Human seasonal influenza viruses




- Influenza virus evolves to avoid human immunity
- Vaccines need frequent updates
Status quo: every few years a big research group publishes a comprehensive analysis.
Can we complement this with automatic always up-to-date analysis?
nextflu.org
joint work with Trevor Bedford & his lab
based on sequences and phenotype provided by the WHO CCs and NICs
nextflu - summary
- relies on data generated in a big coordinated effort by dozens of institutions
- integrates genotypic, phenotypic, and spatial/temporal information
- is always up to date ← possible as data are shared early
- allows interactive exploration of big data sets
- estimates growth of clades → prioritize phenotypic characterization
Real-time molecular epidemiology during outbreaks?
Ebola virus outbreak in West Africa -- Dudas et al, Nature 2017
How do we avoid the long delay?
Bottlenecks:
- sample → DNA to sequence
- sequencing itself not too big a problem
- combining data from different sources
- convincing groups to share and pool their data
- rapid analysis and dissemination of results

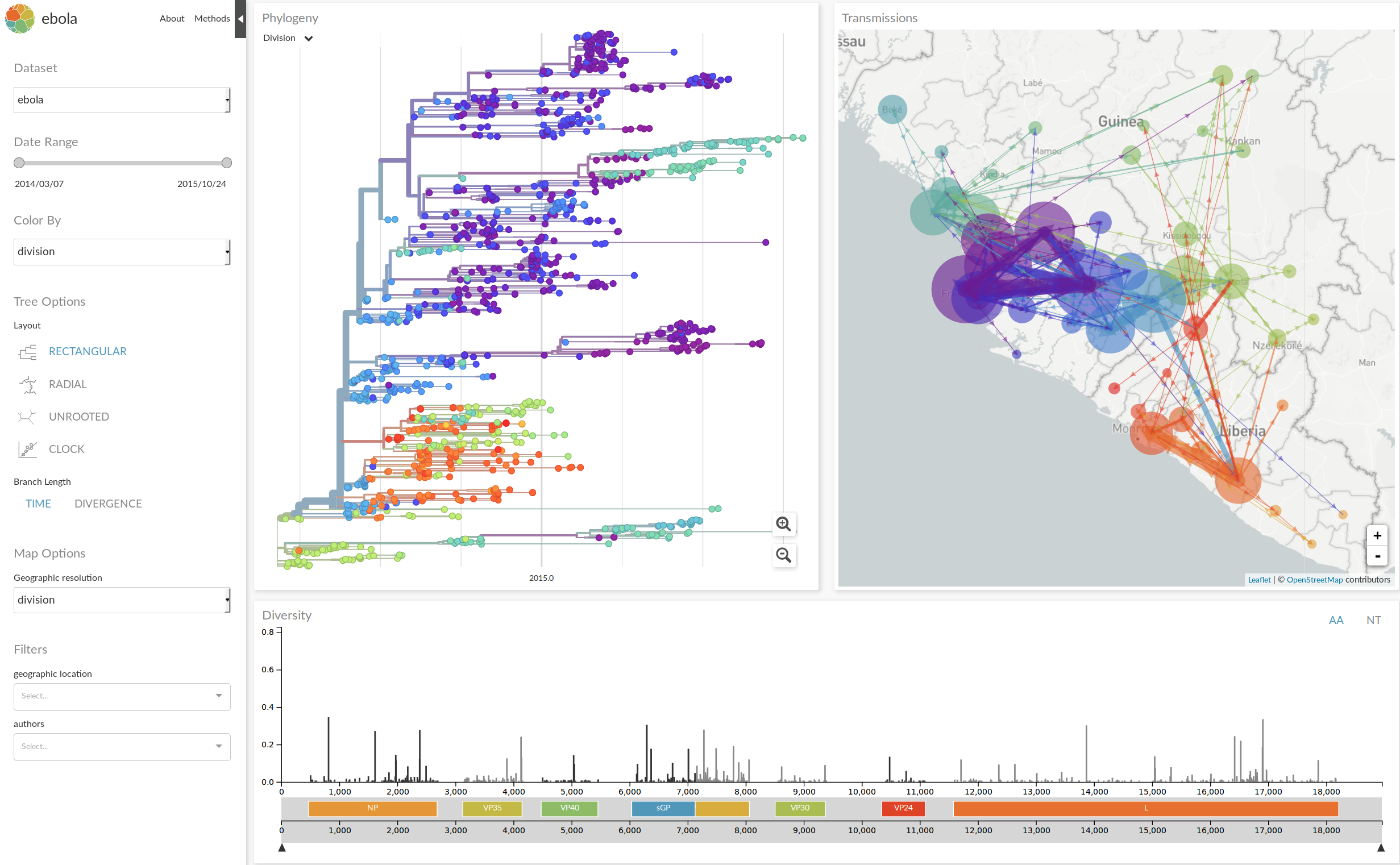
nextstrain.org
- integrate data from many different sources
- analyze those data in near real time
- disseminate results in an intuitive yet informative way
- provide actionable insights
NextStrain architecture
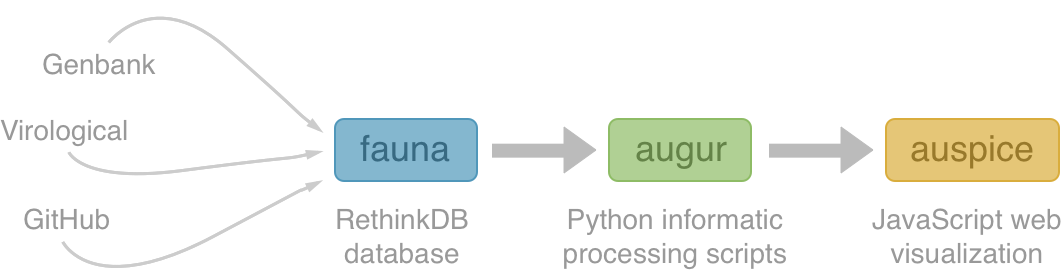
Using treetime to rapidly compute timetrees
nextstrain.org
joint work with Trevor Bedford & his lab
Ian Goodfellow, Matt Cotton, and colleagues sequencing EBOV in West Africa

Beyond tracking: can we predict?
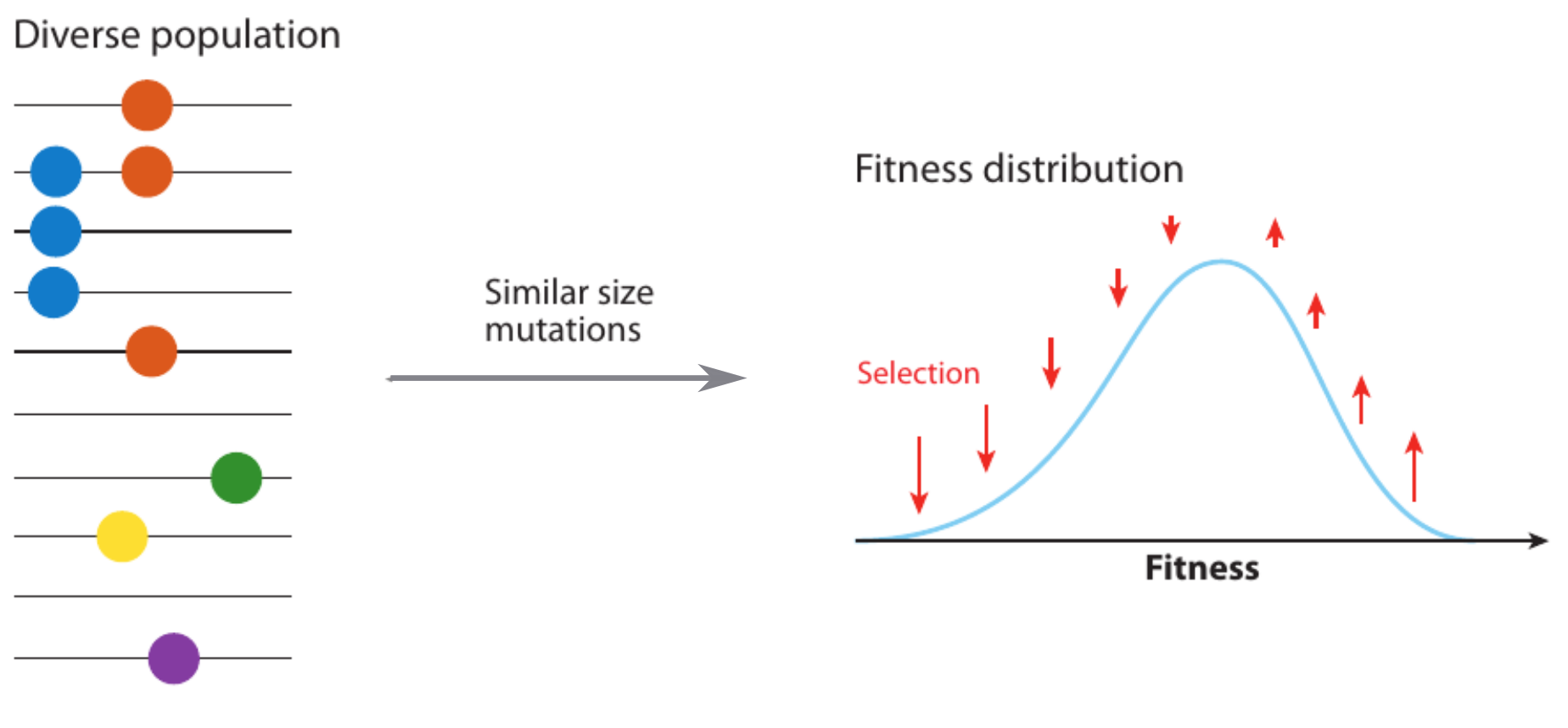
Competition between different viral variants

Bolthausen-Sznitman Coalescent
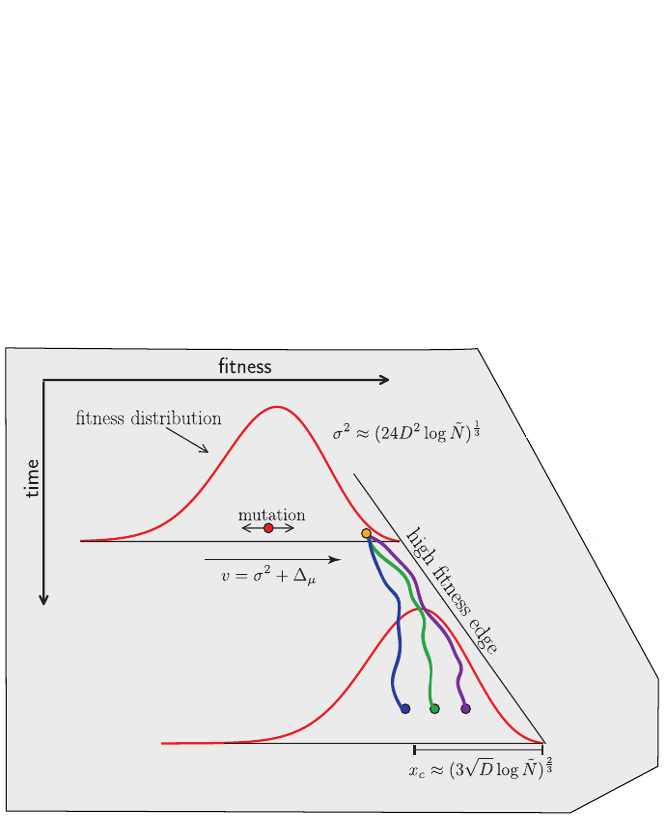
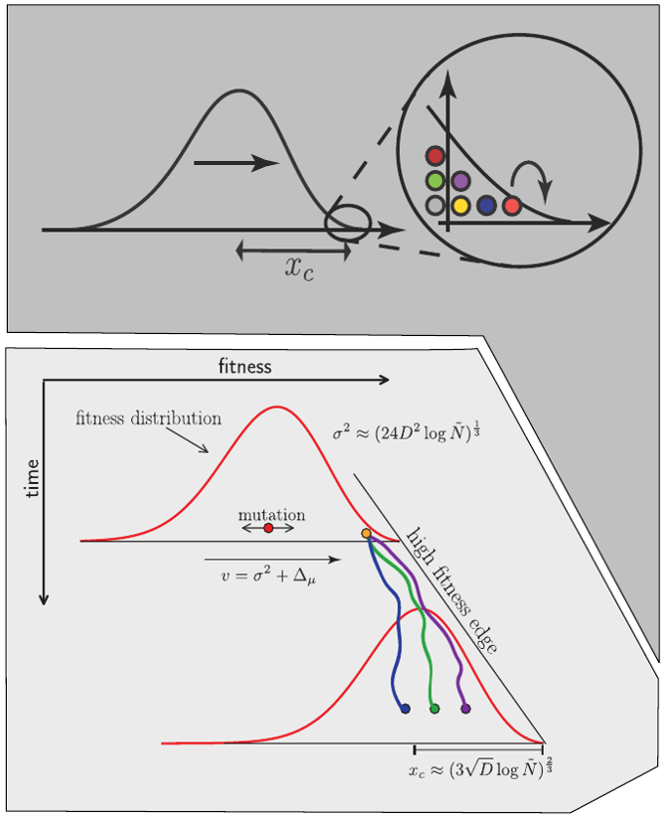
Predicting evolution

Given the branching pattern:
- can we predict fitness?
- pick the closest relative of the future?
Fitness inference from trees
$$P(\mathbf{x}|T) = \frac{1}{Z(T)} p_0(x_0) \prod_{i=0}^{n_{int}} g(x_{i_1}, t_{i_1}| x_i, t_i)g(x_{i_2}, t_{i_2}| x_i, t_i)$$
RN, Russell, Shraiman, eLife, 2014
Prediction of the dominating H3N2 influenza strain
- no influenza specific input
- how can the model be improved? (see model by Luksza & Laessig)
- what other context might this apply?
Challenges and future directions - Routine surveillance
- Nextstrain for every pathogen with surveillance data?
- Automated analysis as a cost-effective way to maximize return
- Animal reservoirs (e.g.~influenza viruses)
- Extension to bacterial pathogens
- Integration with geo-spatial and clinical information
Challenges and future directions - Outbreaks
- Coordination of data sharing
- Alignment of incentives of different stake holders
- Communication of actionable insights to people on the ground


Influenza and Theory acknowledgments




- Boris Shraiman
- Colin Russell
- Trevor Bedford
- Oskar Hallatschek



nextstrain.org
- Trevor Bedford
- Colin Megill
- Pavel Sagulenko
- Sidney Bell
- James Hadfield
- Wei Ding



