Recent progress in predictive modeling
Richard Neher & Trevor Bedford
Biozentrum, University of Basel & Fred Hutch Seattle
slides at neherlab.org/201707_Crick.html
Tools and models
- Tools (nextflu.org / nextstrain.org)
- Flexible display/analysis to HI/FRA data via nextflu
- medium term: move to nextstrain with map
- prediction
- Spotting growing clades: Local branching index
- Spotting growing clades: Frequency trajectories
- Spotting growing clades: antigenic data
nextflu-cdc
nextstrain
Useful additions?
- host age?
- flagging of recurrent mutations?
- glycosylation
- NA, other segments, ...
Prediction: Local Branching Index
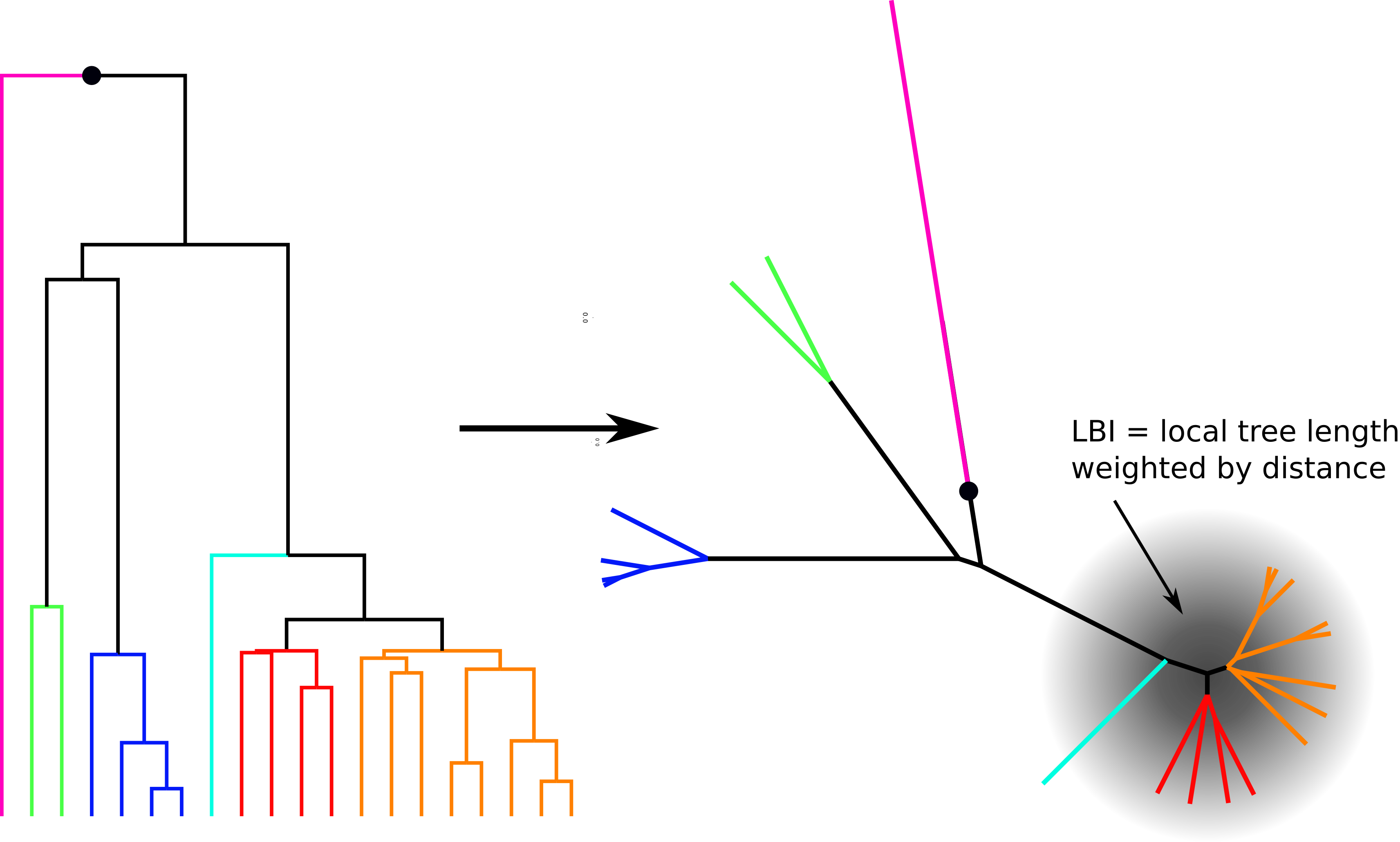
- Single time point
- agnostic/ignorant of biology
- Similar to Luksza/Laessig's phylogenetic component
Prediction: Frequencies
- With time resolved data, more accurate extrapolation should be possible
- Nextflu estimates smoothed frequency trajectories
- Region specific frequencies could use all available data
- Concerns regarding oversampling of certains regions/cities
Prediction: antigenic data
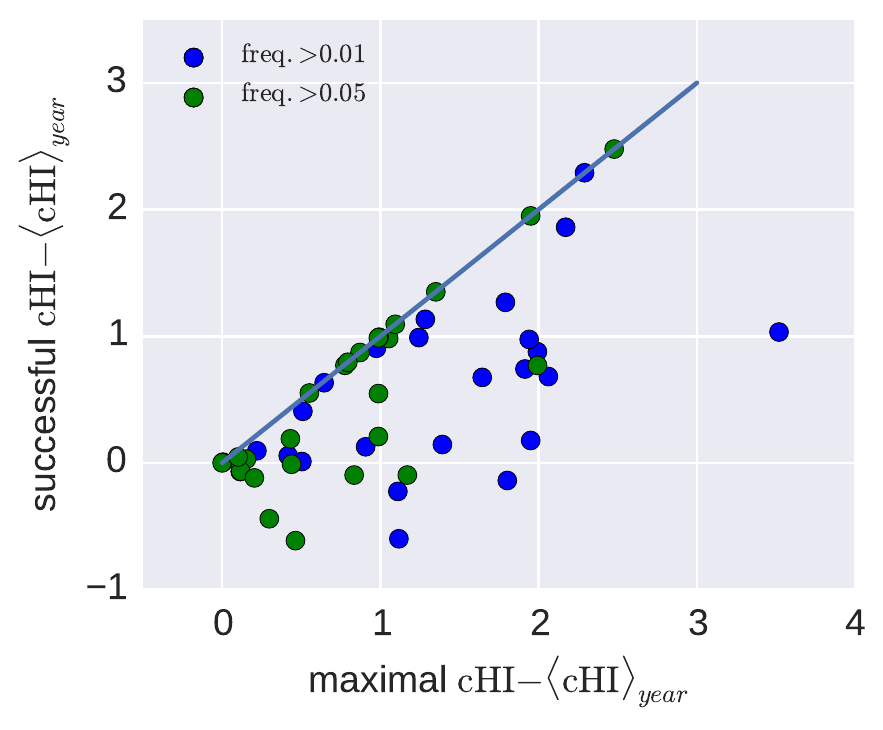
- Antigenic advance is associated with success
- Small advanced clades nevertheless die out
Challenges
- Combining different signals requires parameters
- We have only about 20y to train models on
- Overfitting is a serious concern
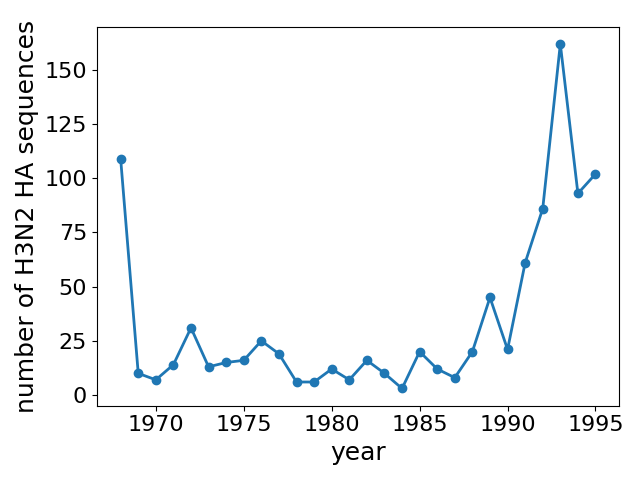
- Generate more data: Sequence 200 viruses per year from 1970 - 1995??