Tracking and predicting evolution and spread of RNA viruses
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/201709_USB_DoKo.html
Evolution of RNA viruses
- Constant struggle to adapt to changing environments and host immunity
- Mutation rates of about 0.00001/site and replication
- Large viral populations explore every single mutation every day
- Most mutations are detrimental, but some persist
- Mutations can act like an approximate clock
Phylogenetic analysis
Phylogenetic analysis
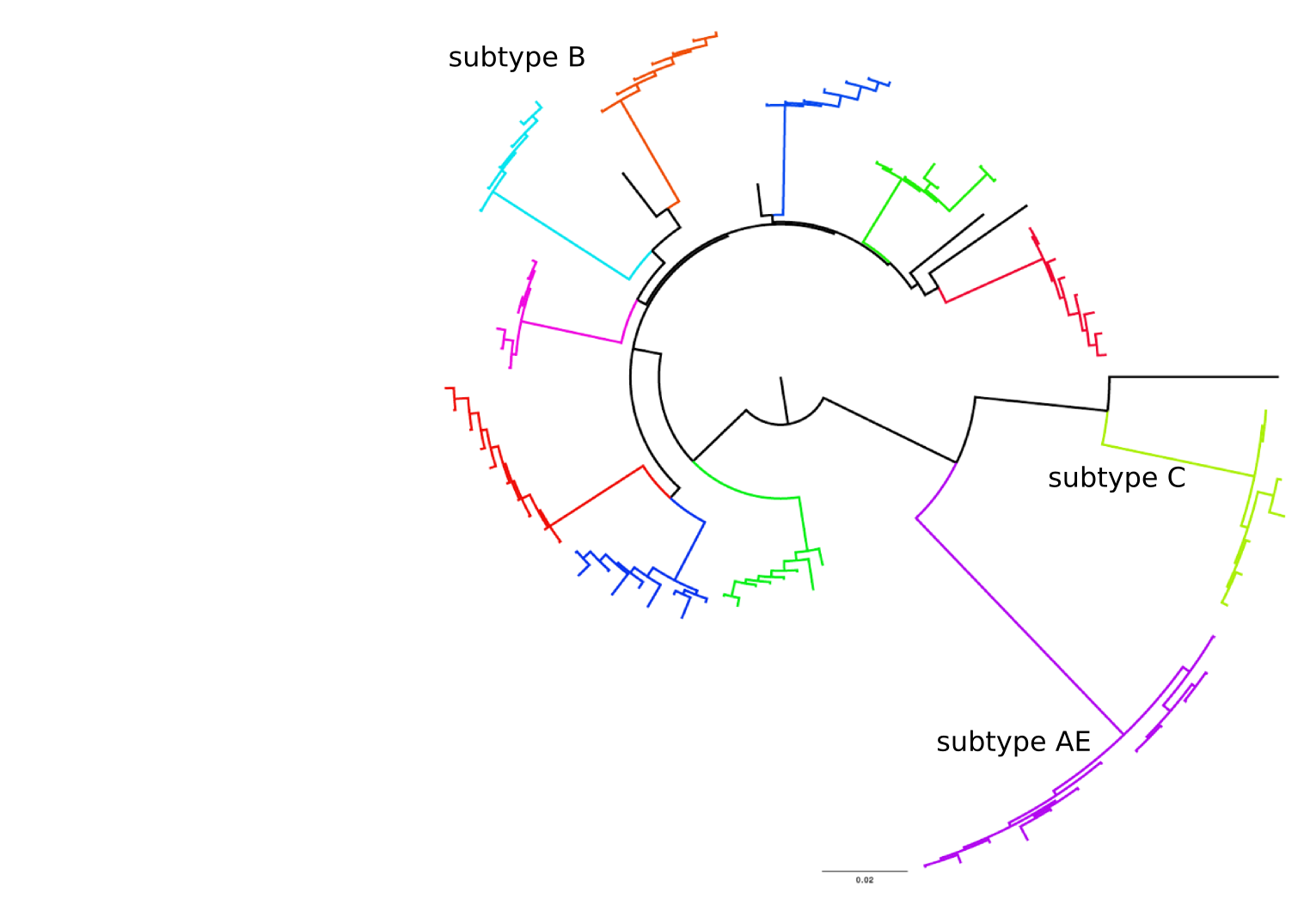
Evolution of HIV
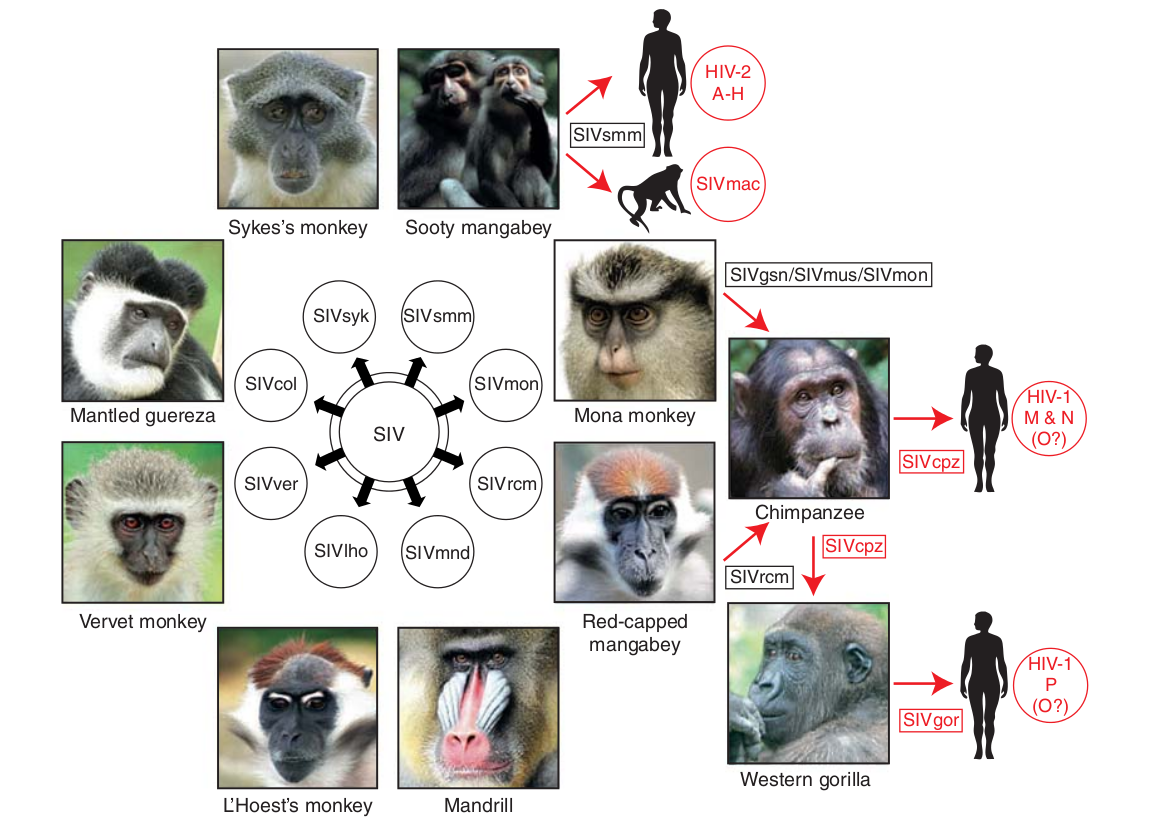
- Chimp → human transmission around 1900 gave rise to HIV-1 group M
- ~100 million infected people since
- subtypes differ at 10-20% of their genome
- HIV-1 evolves ~0.1% per year
HIV infection
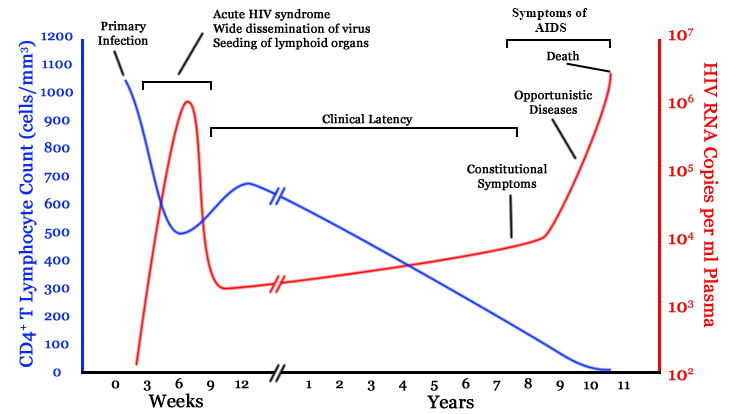
- $10^8$ cells are infected every day
- the virus repeatedly escapes immune recognition
- integrates into T-cells as
latent provirus
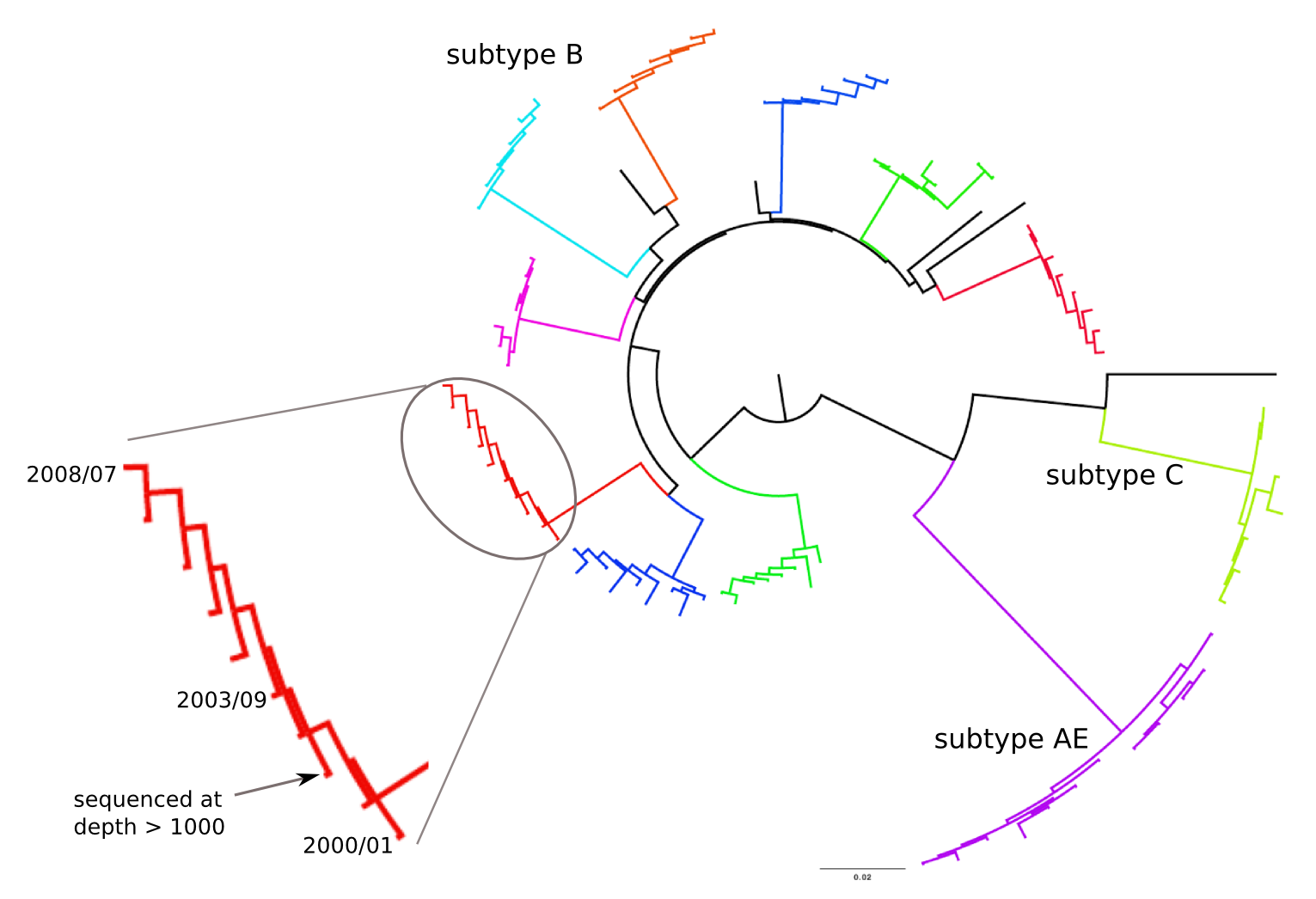
HIV-1 evolution within one individual
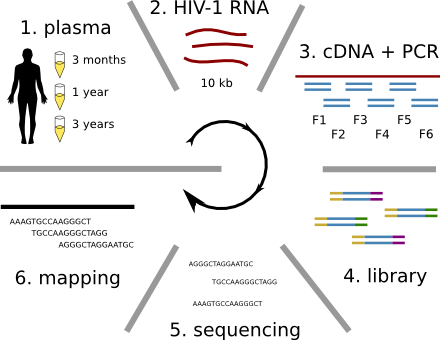

HIV-1 sequencing before and after therapy
 Zanini et al, eLife, 2015;
Brodin et al, eLife, 2016.
Collaboration with the group of Jan Albert
Zanini et al, eLife, 2015;
Brodin et al, eLife, 2016.
Collaboration with the group of Jan Albert
Population sequencing to track all mutations above 1%
- diverge at 0.1-1% per year
- almost full genomes coverage in 10 patients
- full data set at hiv.tuebingen.mpg.de
Diversity and rates of change
- envelope changes fastest, enzymes slowest
- identical rate of synonymous evolution
- diversity saturates where evolution is fast
- synonymous mutations stay at low frequency
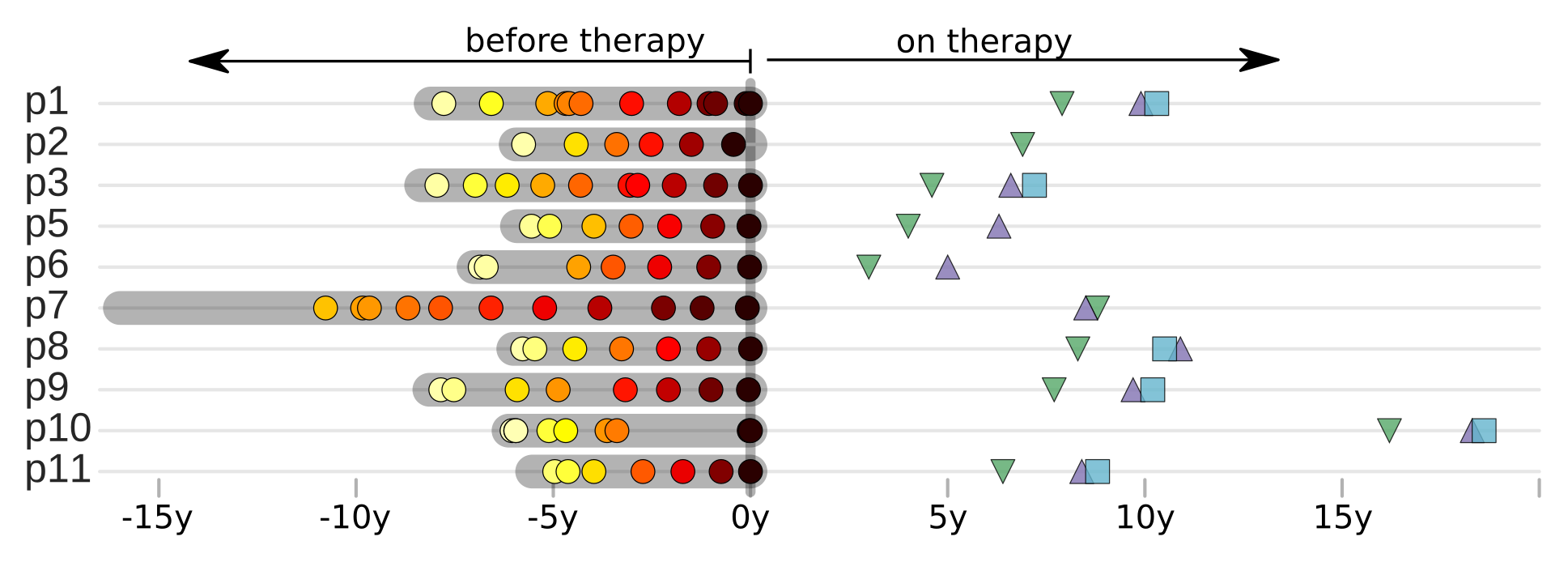
Does HIV evolve during therapy?
 Brodin et al, eLife, 2016
Brodin et al, eLife, 2016
No evidence of ongoing replication
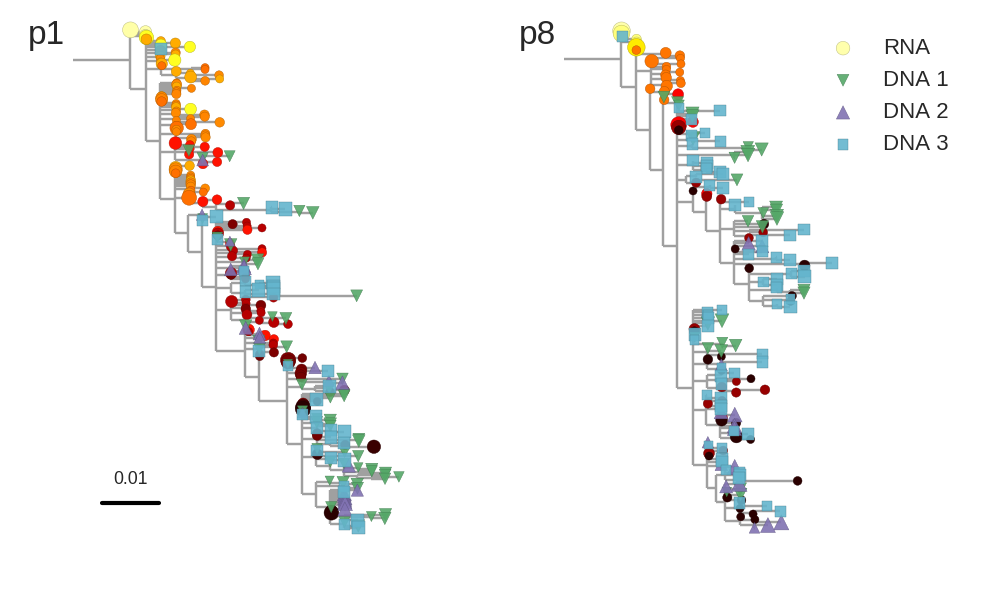
No evidence of ongoing replication
Sequences record the spread of pathogens




The resolution is limited by the number of mutations!
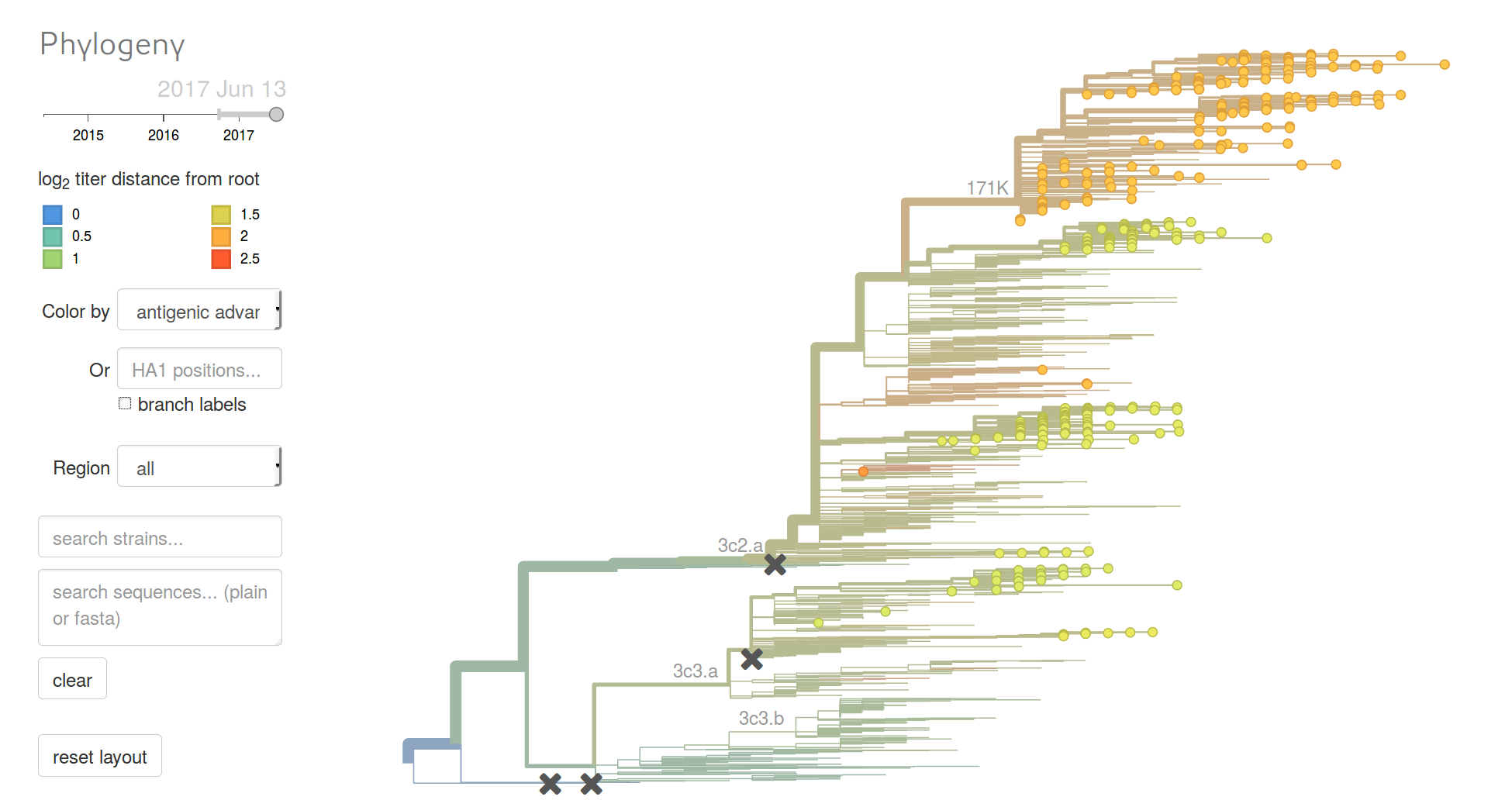
Human seasonal influenza viruses




- Influenza virus evolves to avoid human immunity
- Vaccines need frequent updates
nextflu.org
joint work with Trevor Bedford & his lab
based on sequences and phenotype provided by the WHO CCs and NICs
Beyond tracking: can we predict?
Competition between different viral variants
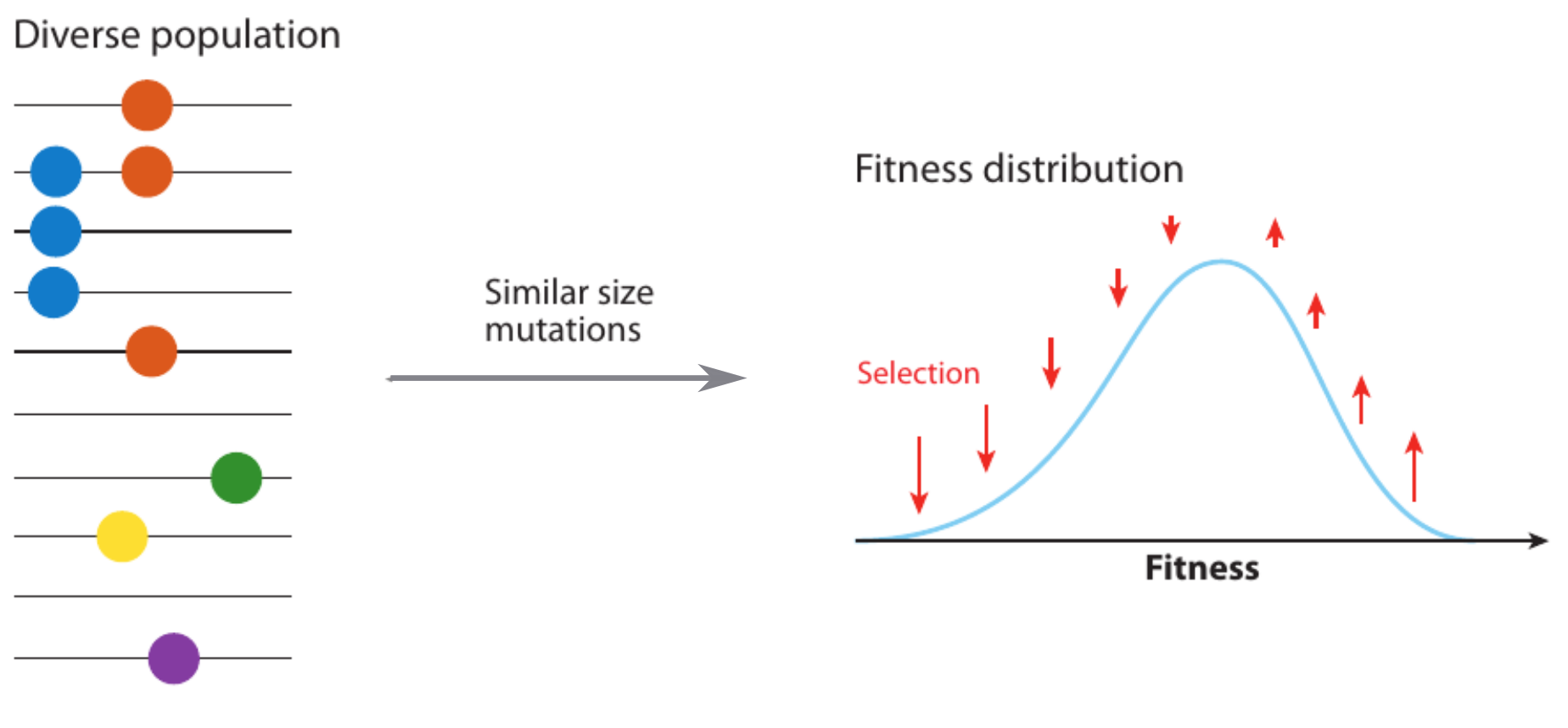
Predicting evolution

Given the branching pattern:
- can we predict fitness?
- pick the closest relative of the future?
Fitness inference from trees
$$P(\mathbf{x}|T) = \frac{1}{Z(T)} p_0(x_0) \prod_{i=0}^{n_{int}} g(x_{i_1}, t_{i_1}| x_i, t_i)g(x_{i_2}, t_{i_2}| x_i, t_i)$$
RN, Russell, Shraiman, eLife, 2014
Prediction of the dominating H3N2 influenza strain
- no influenza specific input
- how can the model be improved? (see model by Luksza & Laessig)
- what other context might this apply?
nextstrain.org
- integrate data from many different sources
- analyze those data in near real time
- disseminate results in an intuitive yet informative way
- provide actionable insights
Acknowledgments
- Fabio Zanini
- Jan Albert
- Johanna Brodin
- Christa Lanz
- Göran Bratt
- Lina Thebo
- Vadim Puller



Influenza and Theory acknowledgments




- Boris Shraiman
- Colin Russell
- Trevor Bedford
- Oskar Hallatschek



nextstrain.org
- Trevor Bedford
- Colin Megill
- Pavel Sagulenko
- Sidney Bell
- James Hadfield
- Wei Ding



