Antigenic evolution of seasonal influenza viruses
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/201710_PCCMI.html
Human seasonal influenza viruses




- Influenza viruses evolve to avoid human immunity
- Vaccines need frequent updates

nextflu.org
joint work with Trevor Bedford & his lab
code at github.com/blab/nextflu
Hemagglutination Inhibition assays
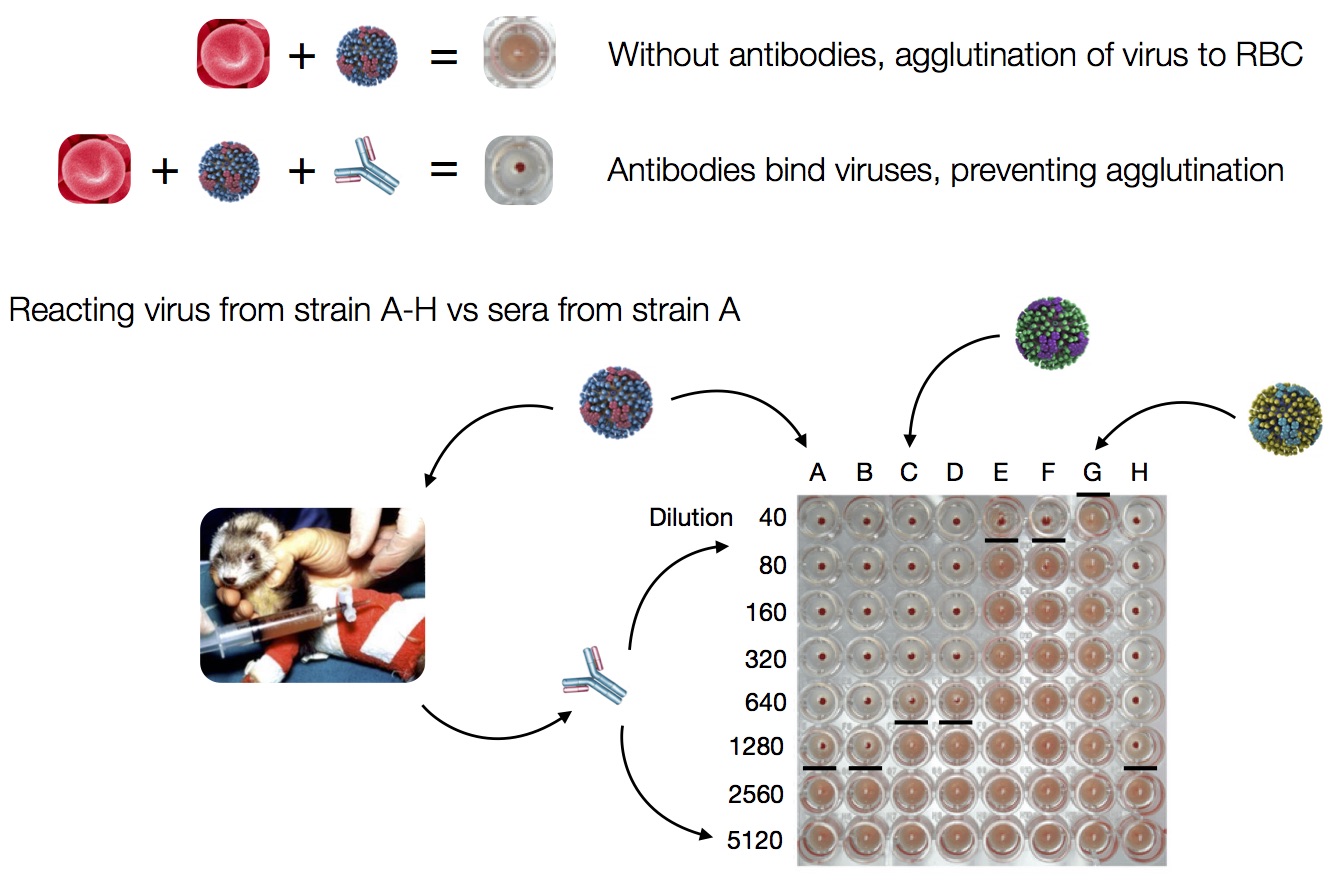
HI data sets
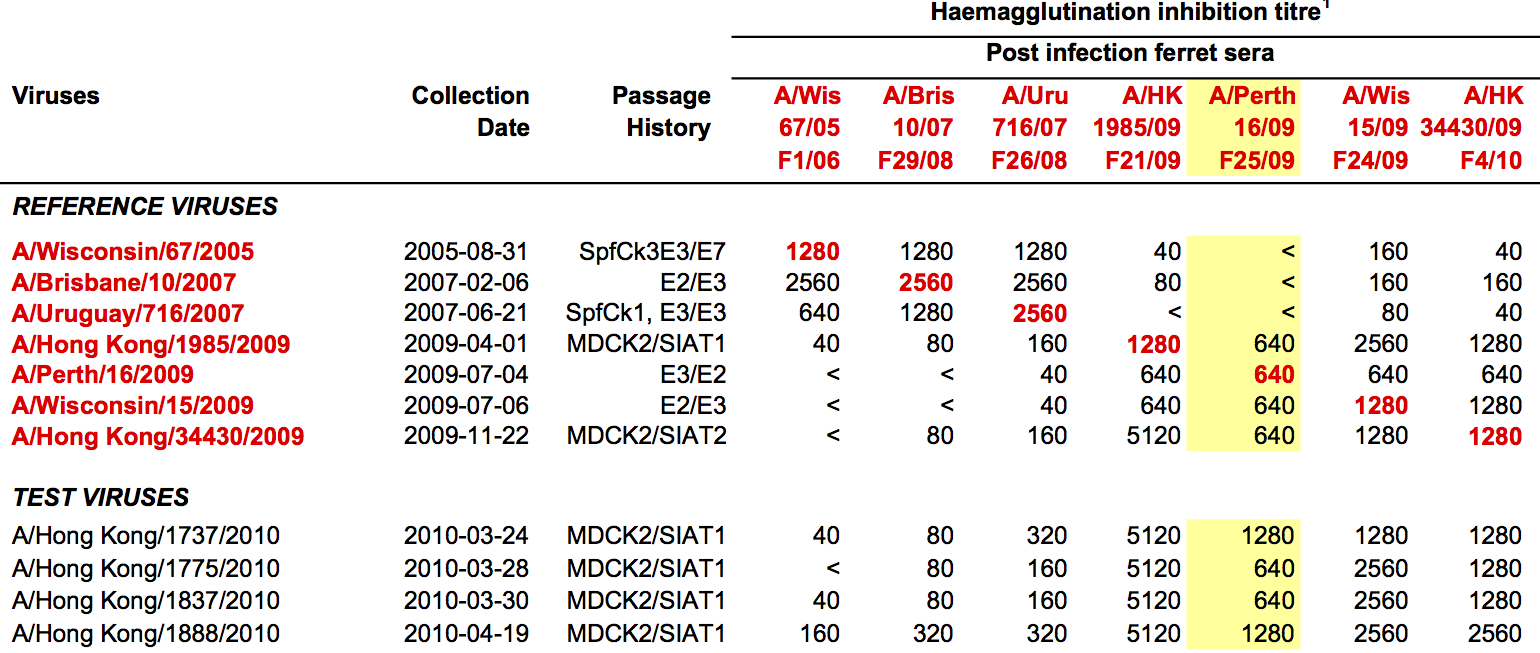
- Long list of distances between sera and viruses
- Tables are sparse, only close by pairs
- Structure of space is not immediately clear
- MDS in 2 or 3 dimensions
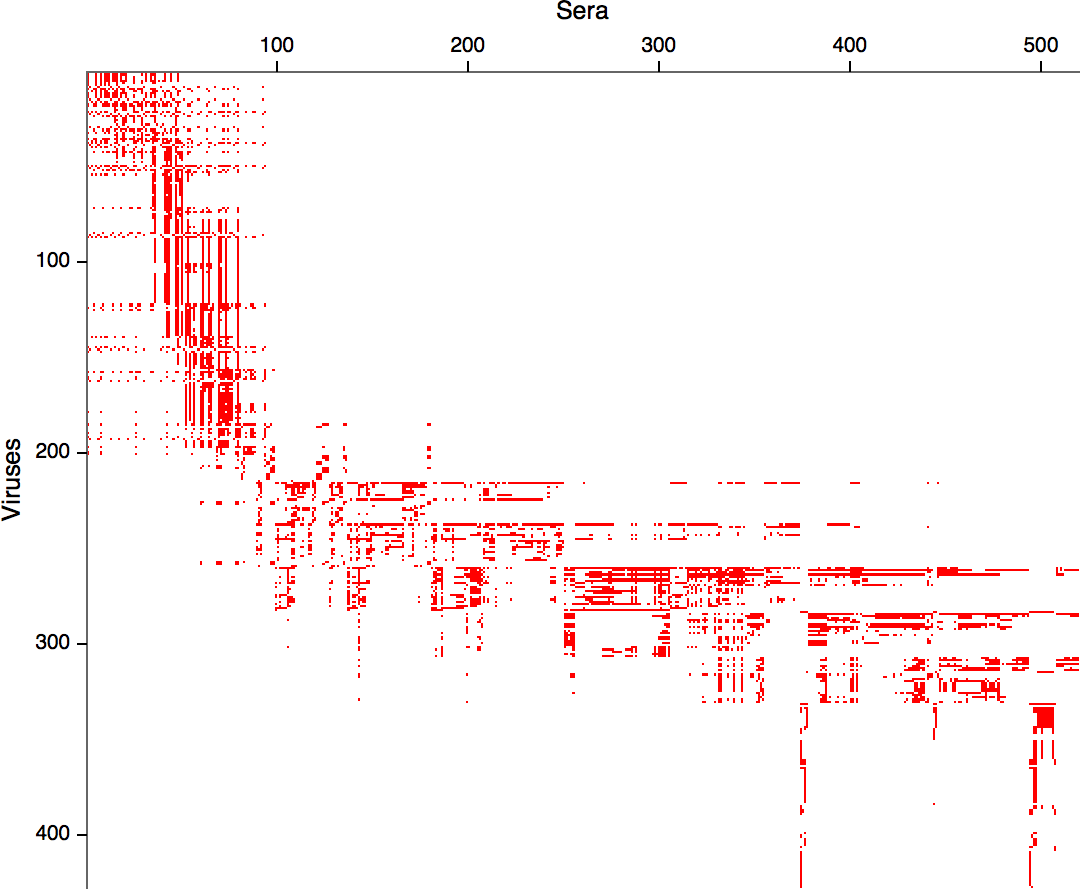
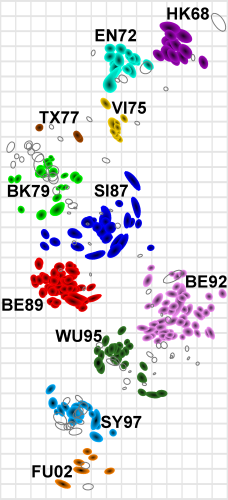 Smith et al, Science 2002
Smith et al, Science 2002
Integrating antigenic and molecular evolution
- $H_{a\beta} = v_a + p_\beta + \sum_{i\in (a,b)} d_i$
- each branch contributes $d_i$ to antigenic distance
- sparse solution for $d_i$ through $l_1$ regularization
- related model where $d_i$ are associated with substitutions
Integrating antigenic and molecular evolution
- MDS: $(d+1)$ parameters per virus
- Tree model: $2$ parameters per virus
- Sparse solution
→ identify branches or substitutions that cause titer drop
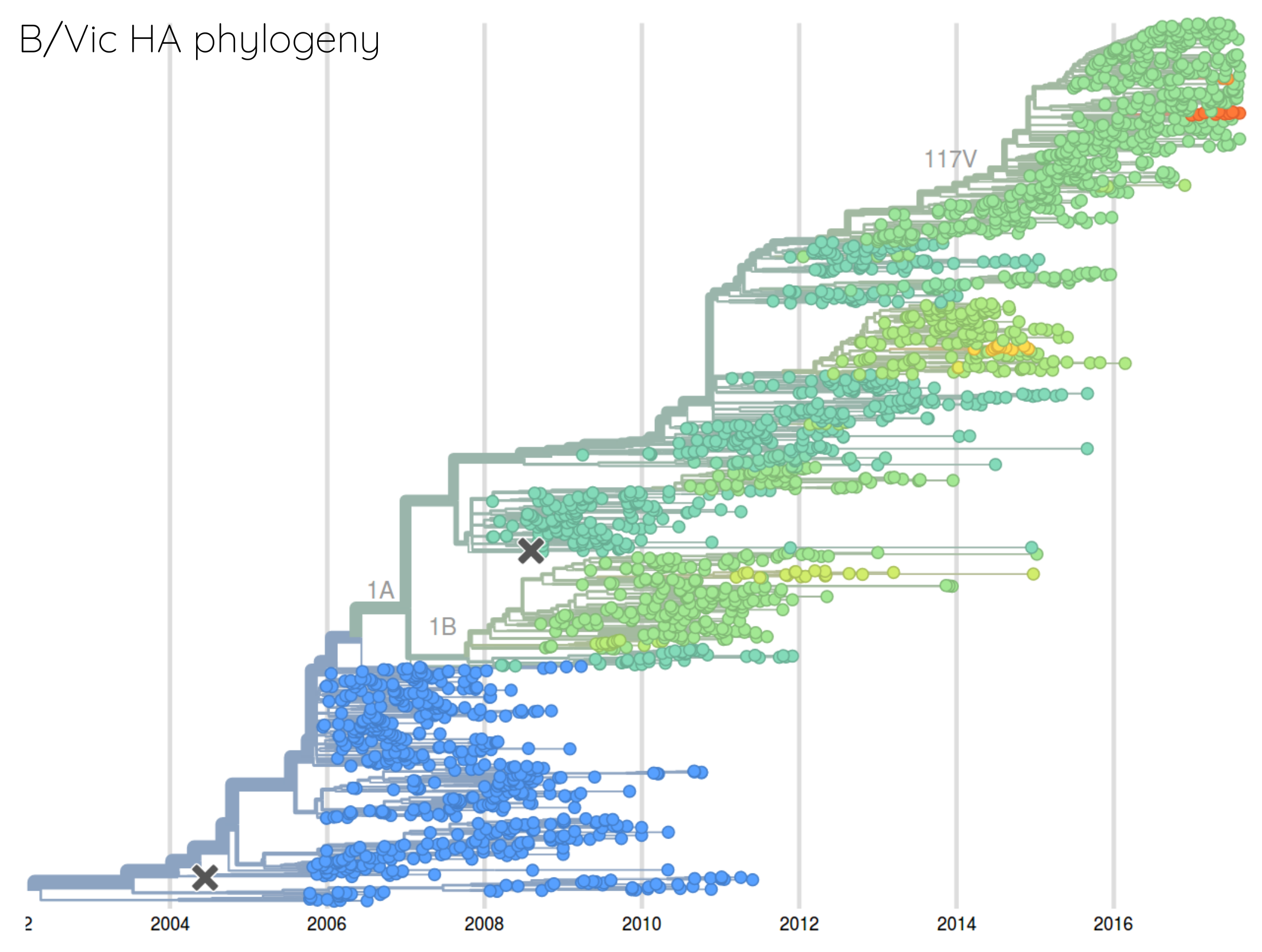
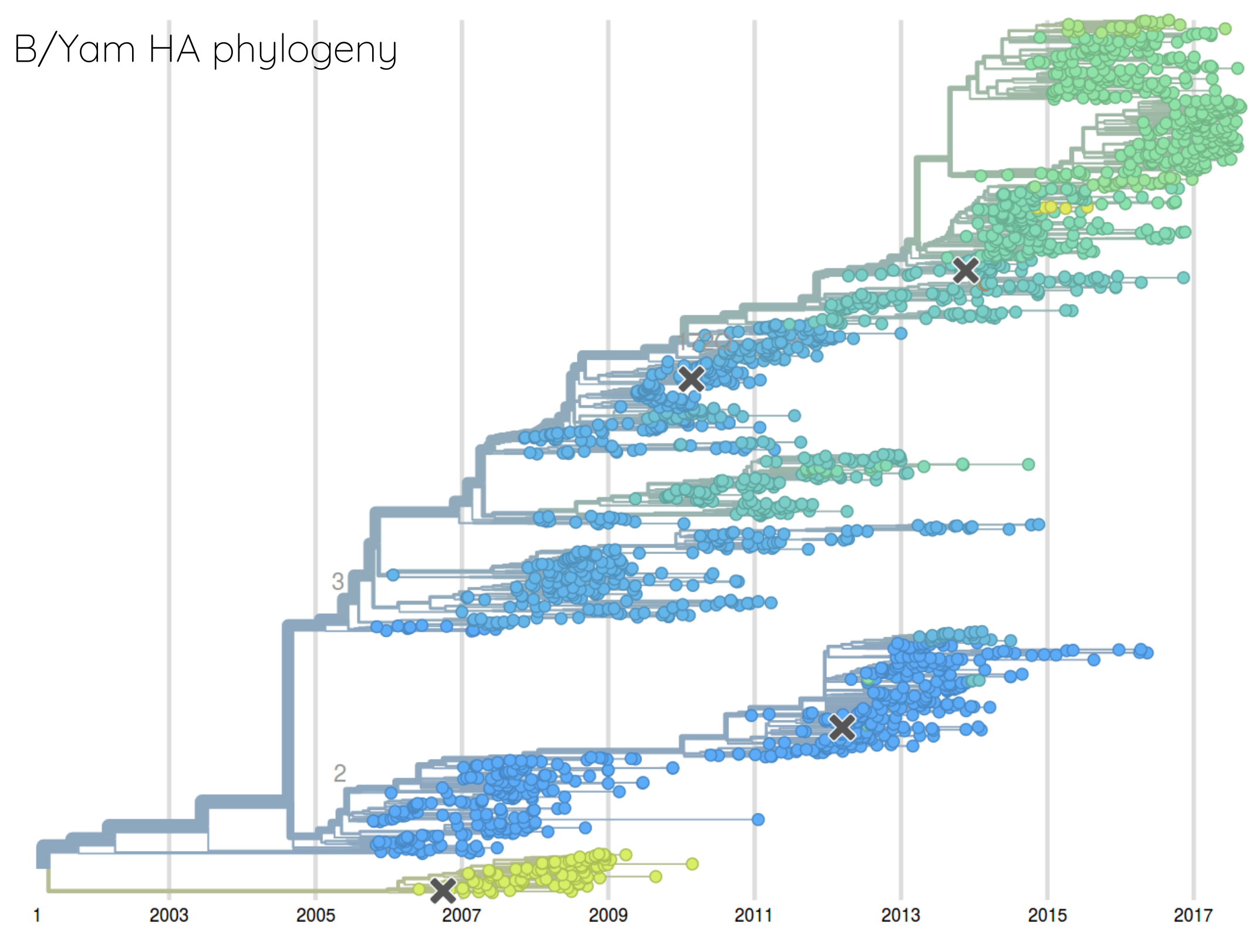
Are antigenic distances tree-like?
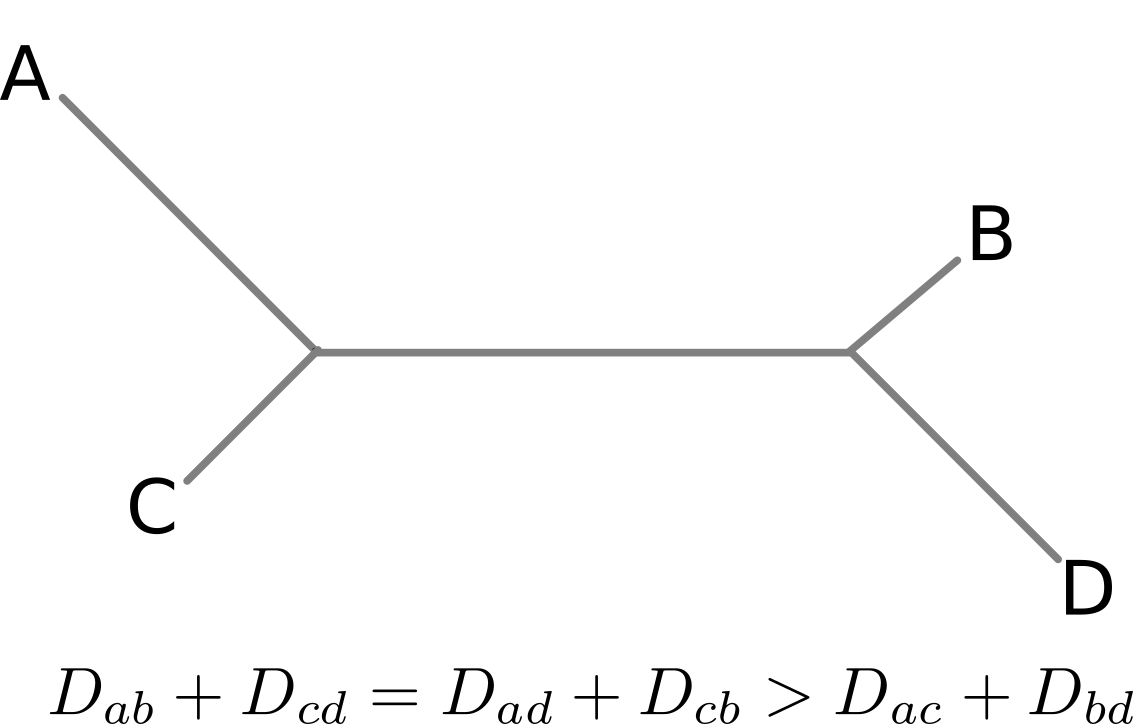
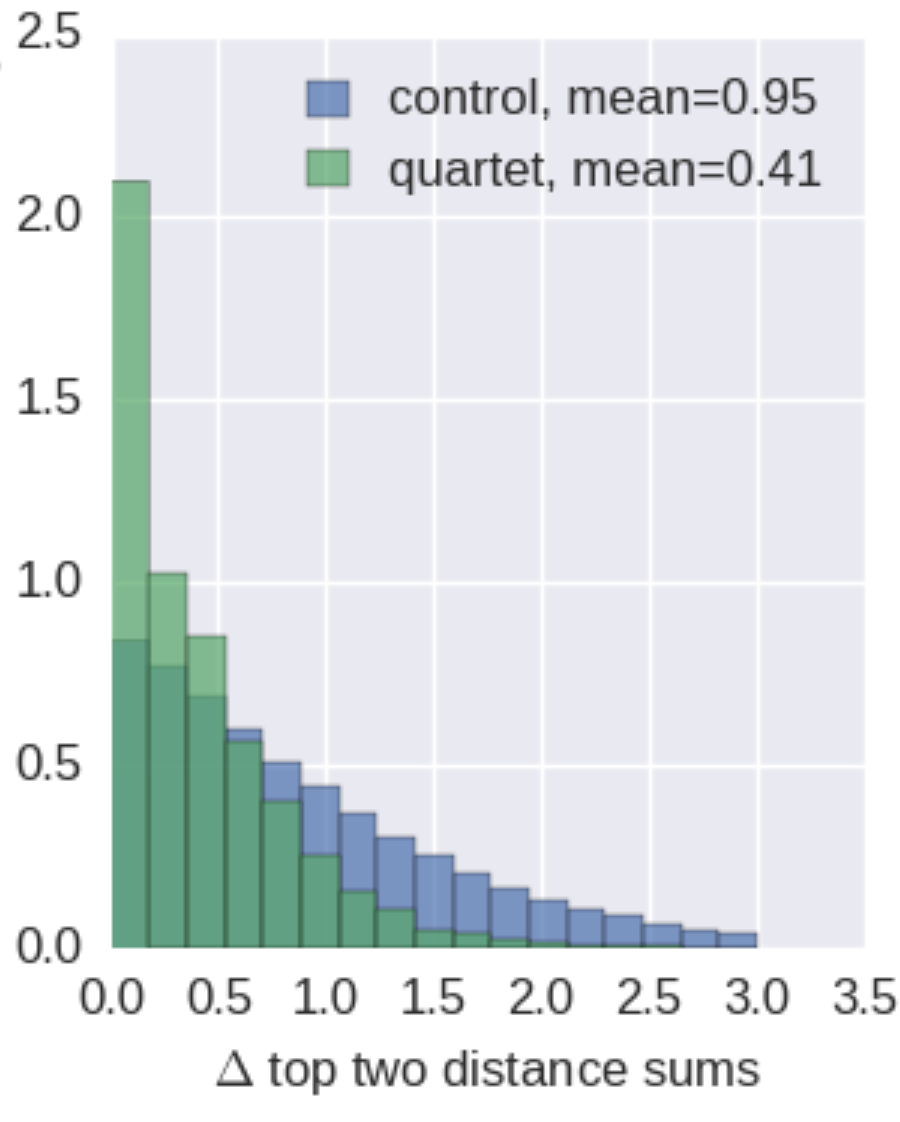
Rate of antigenic evolution
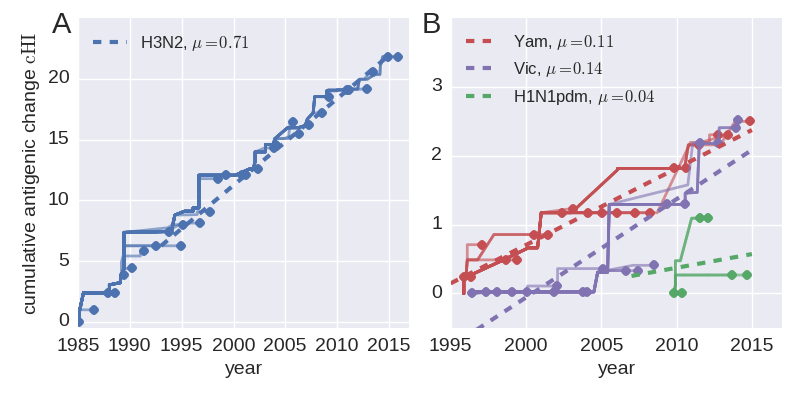
- Cumulative antigenic evolution since the root: $\sum_i d_i$
- A/H3N2 evolves faster antigenically
- A/H3N2 has a more rapid population turn-over
- Proportion of children is high in B vs A/H3N2 infections
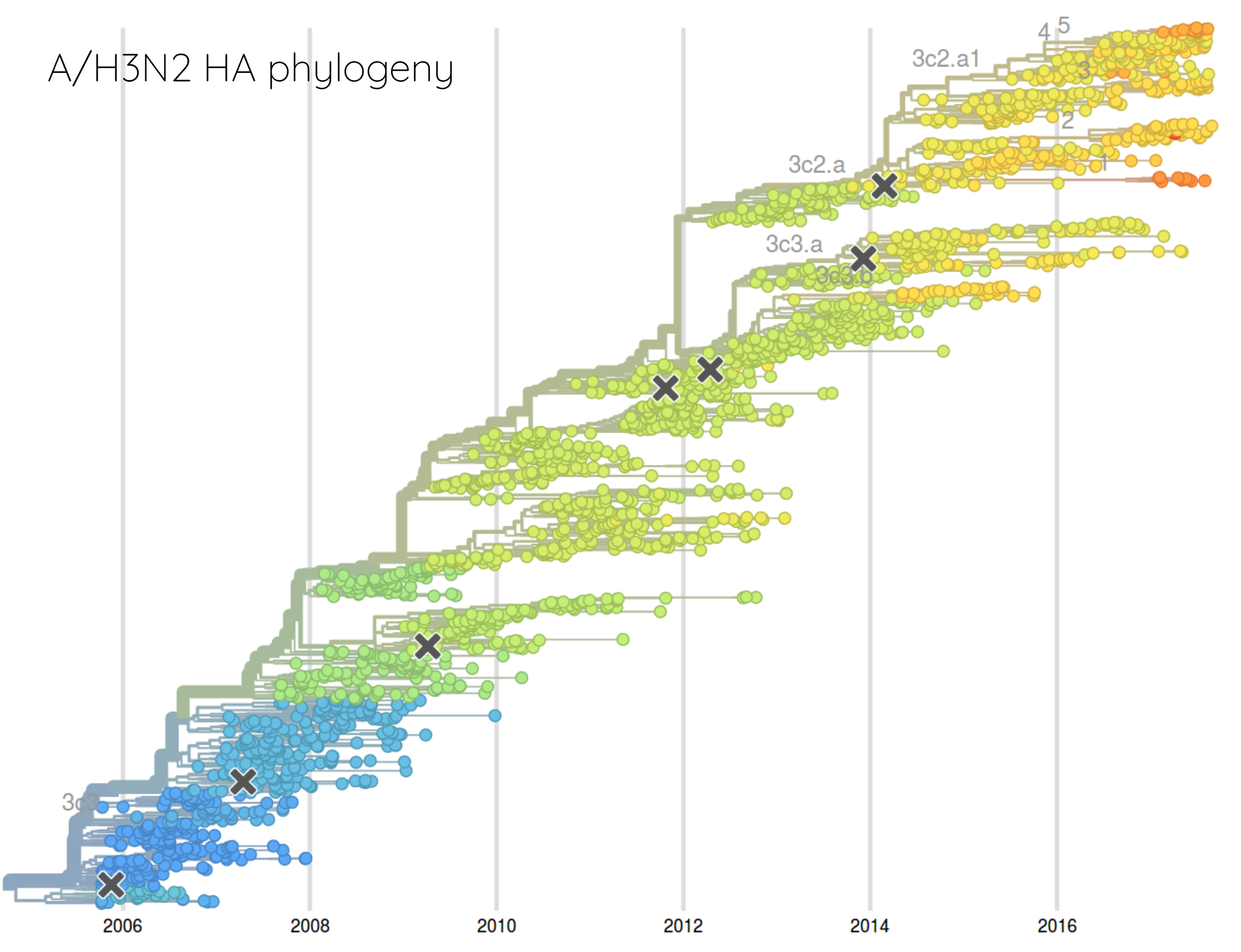
How many sites are involved?

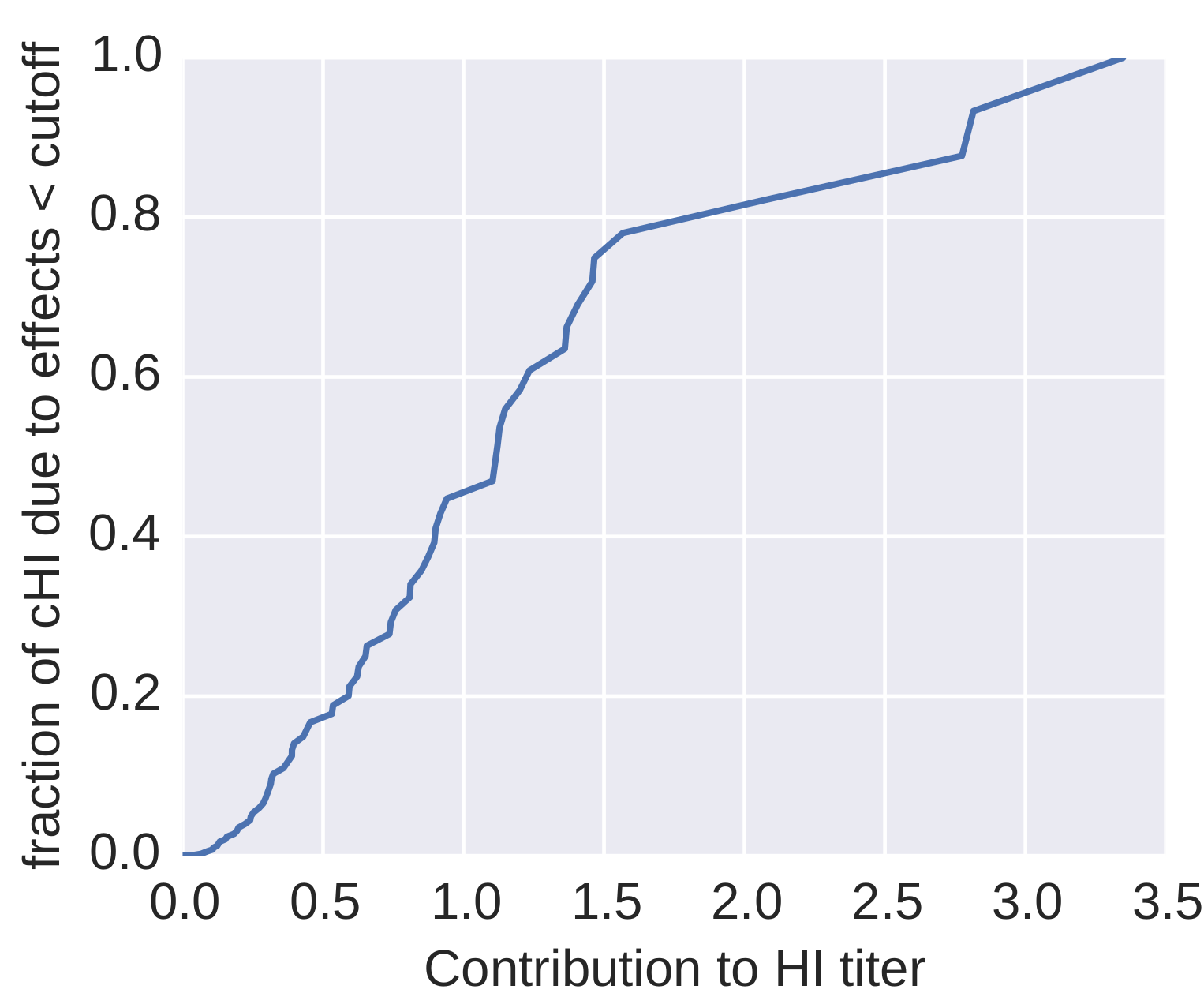
| Mutation | effect |
| K158N/N189K | 3.64 |
| K158R | 2.31 |
| K189N | 2.18 |
| S157L | 1.29 |
| V186G | 1.25 |
| S193F | 1.2 |
| K140I | 1.1 |
| F159Y | 1.08 |
| K144D | 1.08 |
| K145N | 0.91 |
| S159Y | 0.89 |
| I25V | 0.88 |
| Q1L | 0.85 |
| K145S | 0.85 |
| K144N | 0.85 |
| N145S | 0.85 |
| N8D | 0.73 |
| T212S | 0.69 |
| N188D | 0.65 |
Predicting successful influenza clades
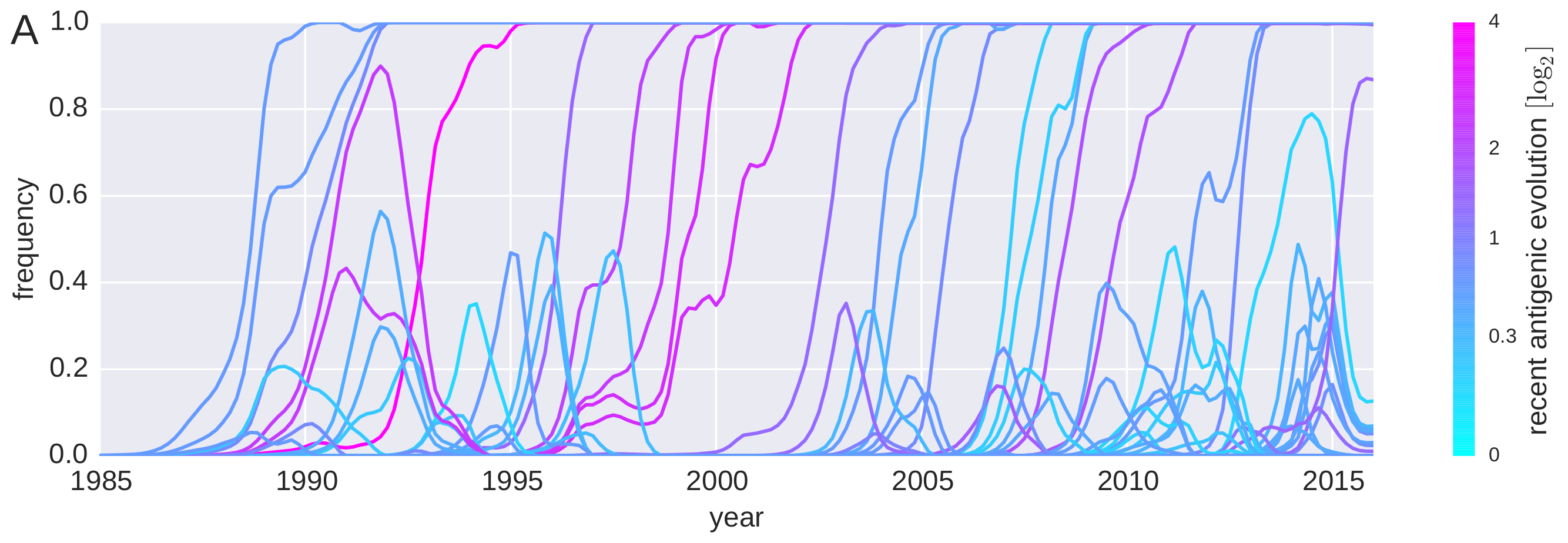
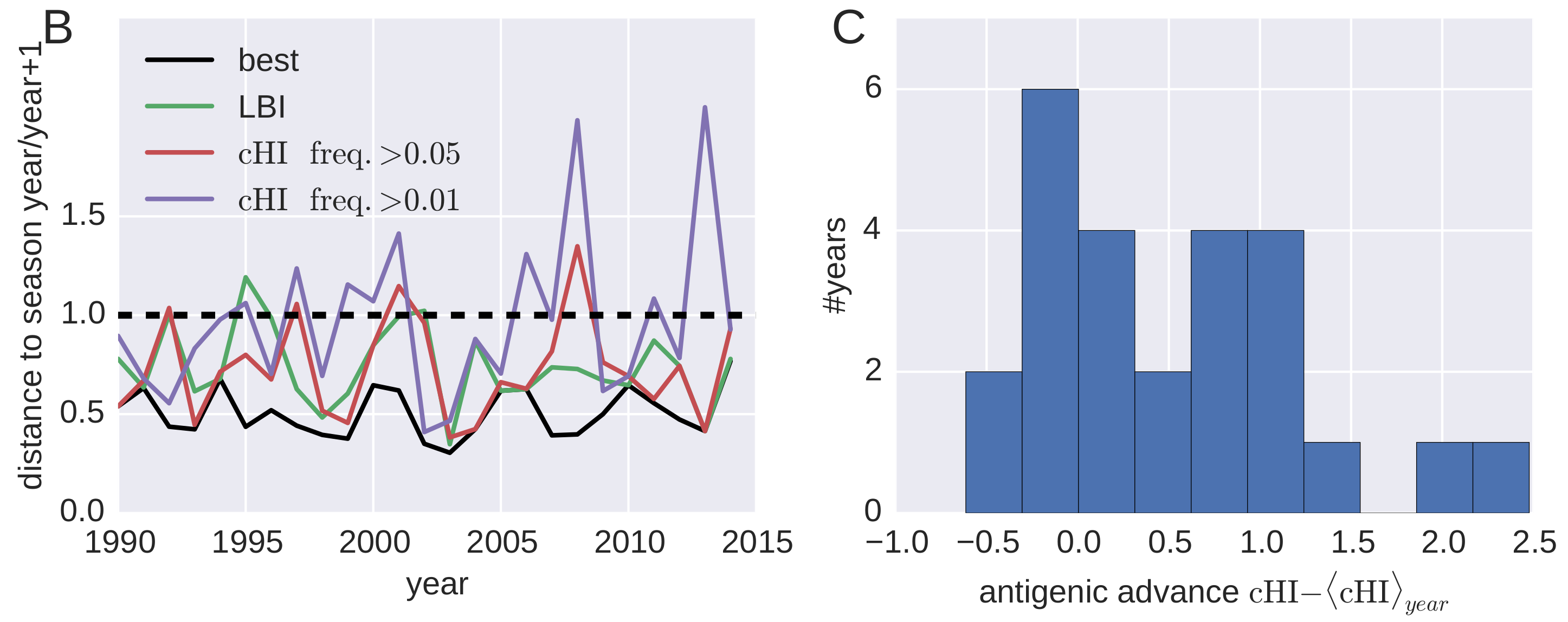
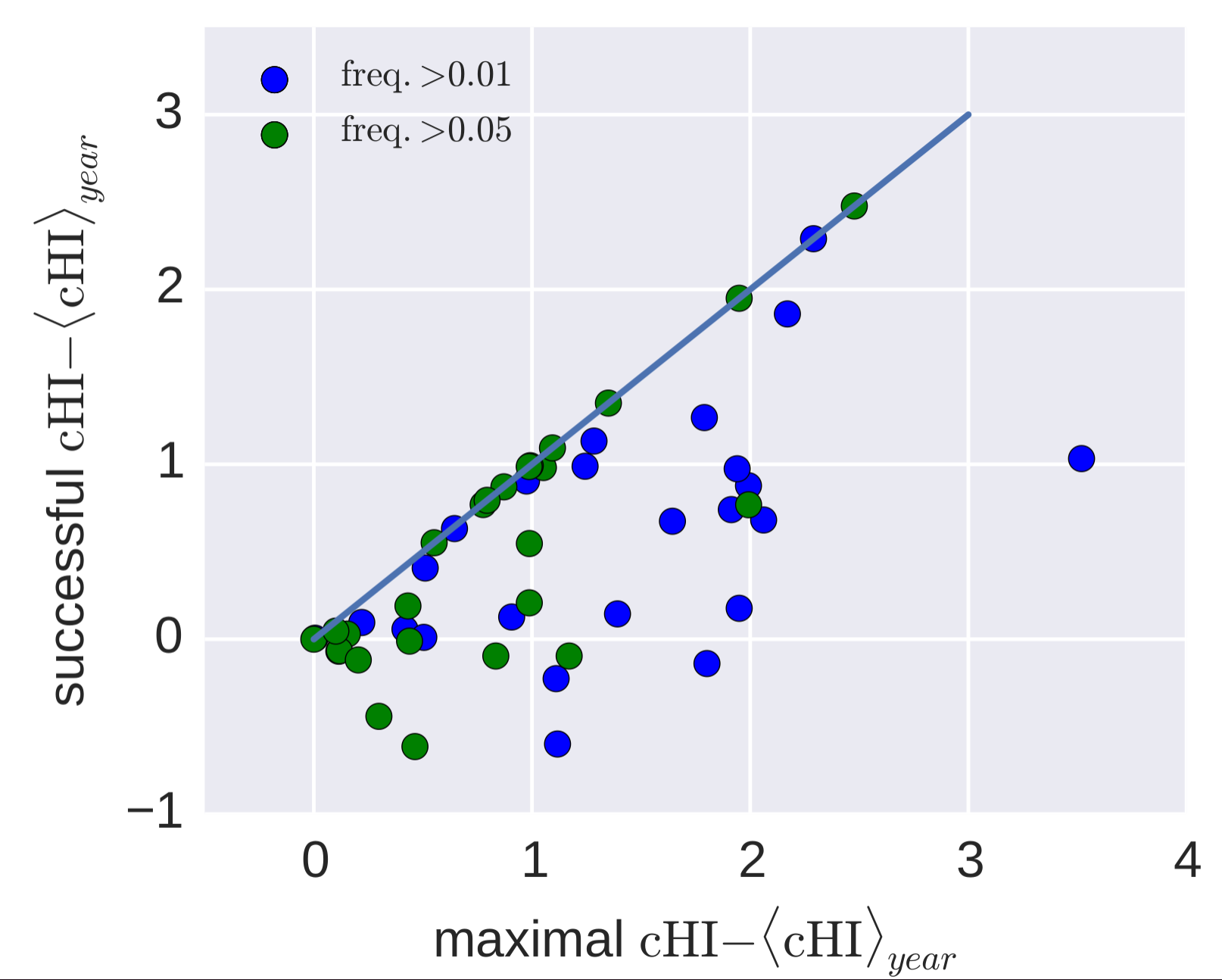
Predicting successful influenza clades
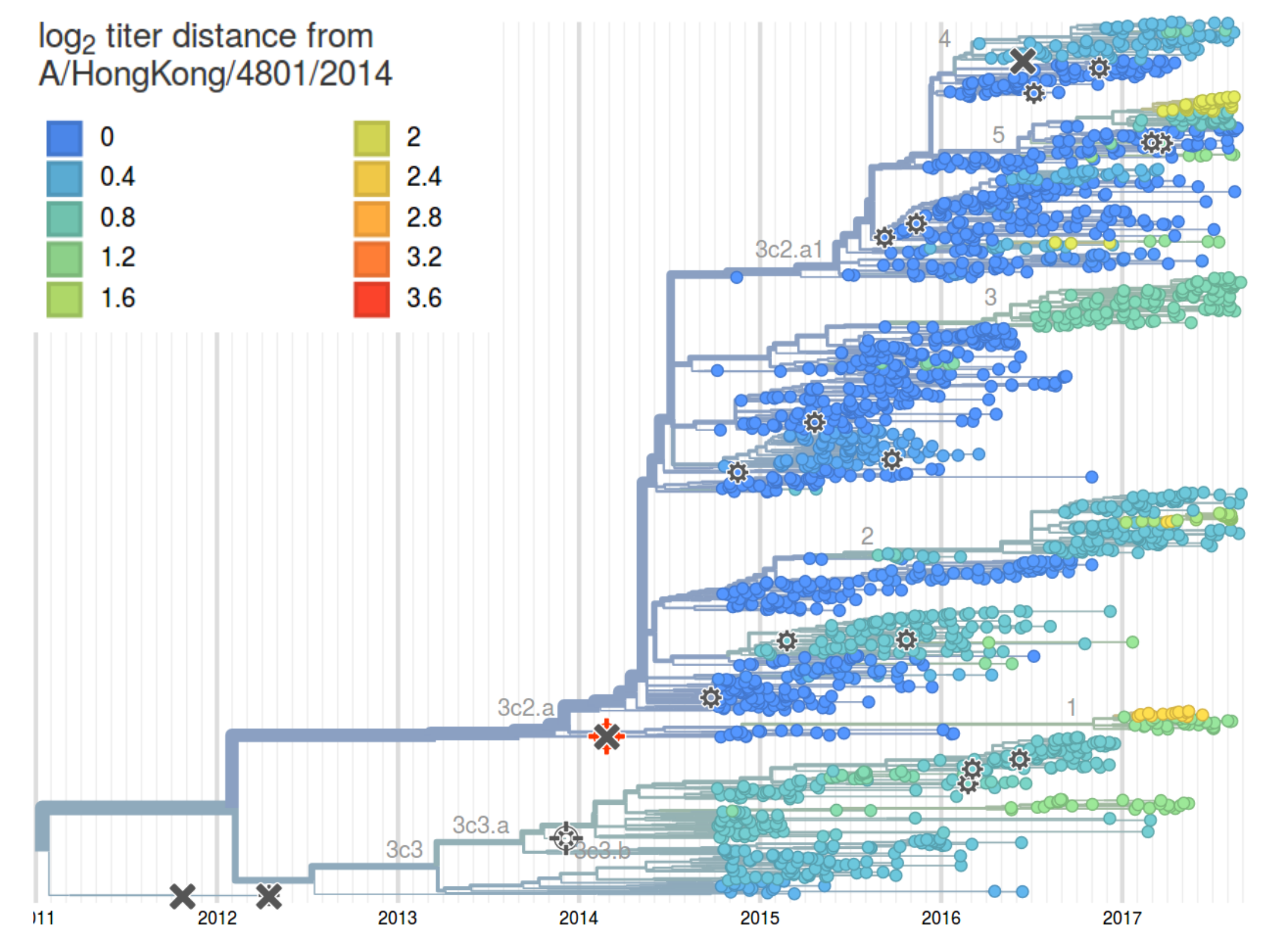
HI distances on the phylogenetic tree
Shortcomings of ferret HI data
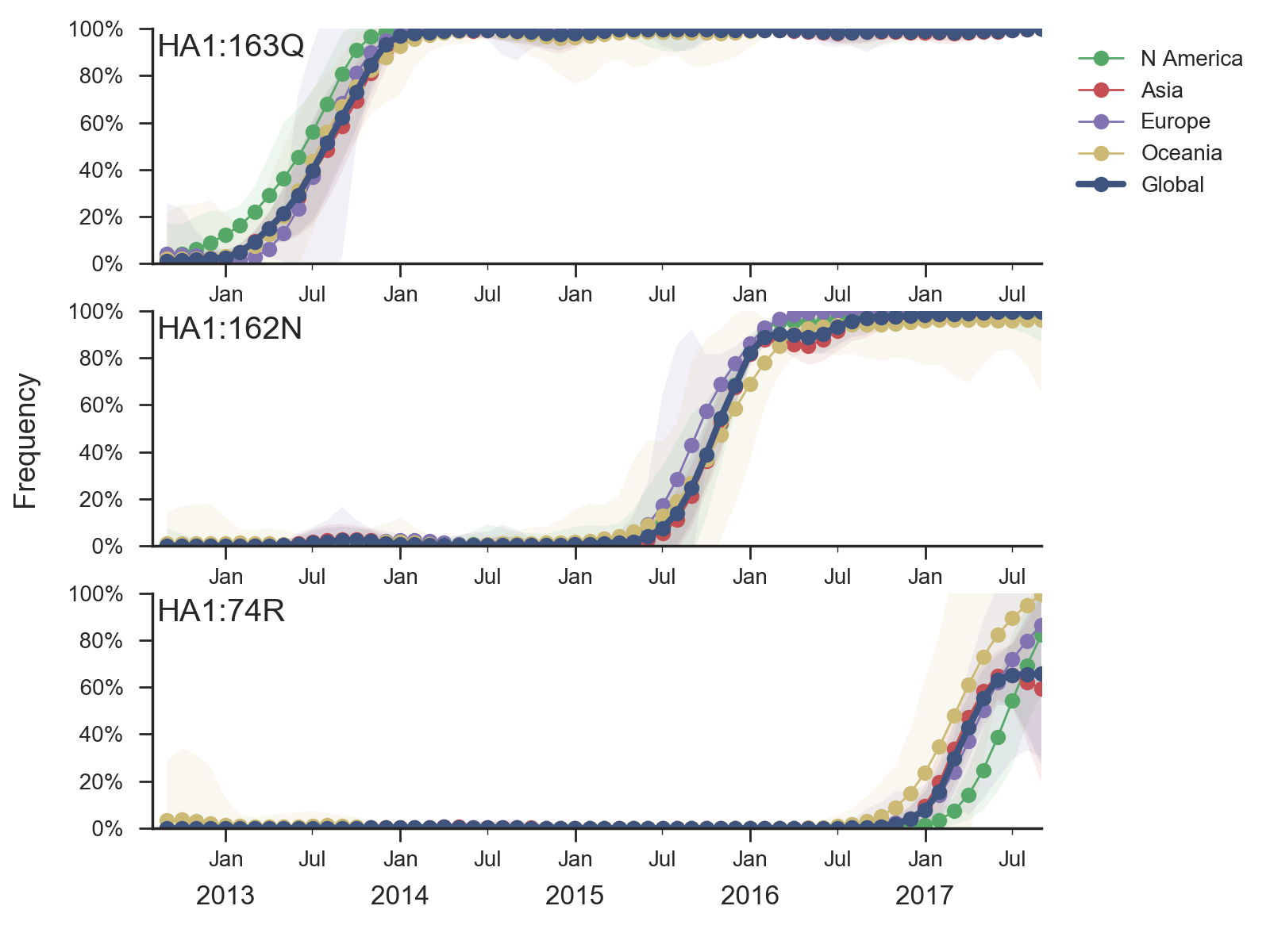
- ferrets are naive -- no complicated history of exposure
- H1N1pdm experienced three recent rapid amino acid replacements
- age specific immuno-dominance caused problems in the 2013-2014 influenza season
Linderman .... Hensley, PNAS 2014
- ABs in people born between 1960 and 1979 target an epitope containing 163K
- Young people target a different epitope -- like ferrets
- The 163K epitope in H1N1pdm is similar to H1N1 viruses circulating 1976-1986
- The vaccine virus was 163K, likely boosted the old response
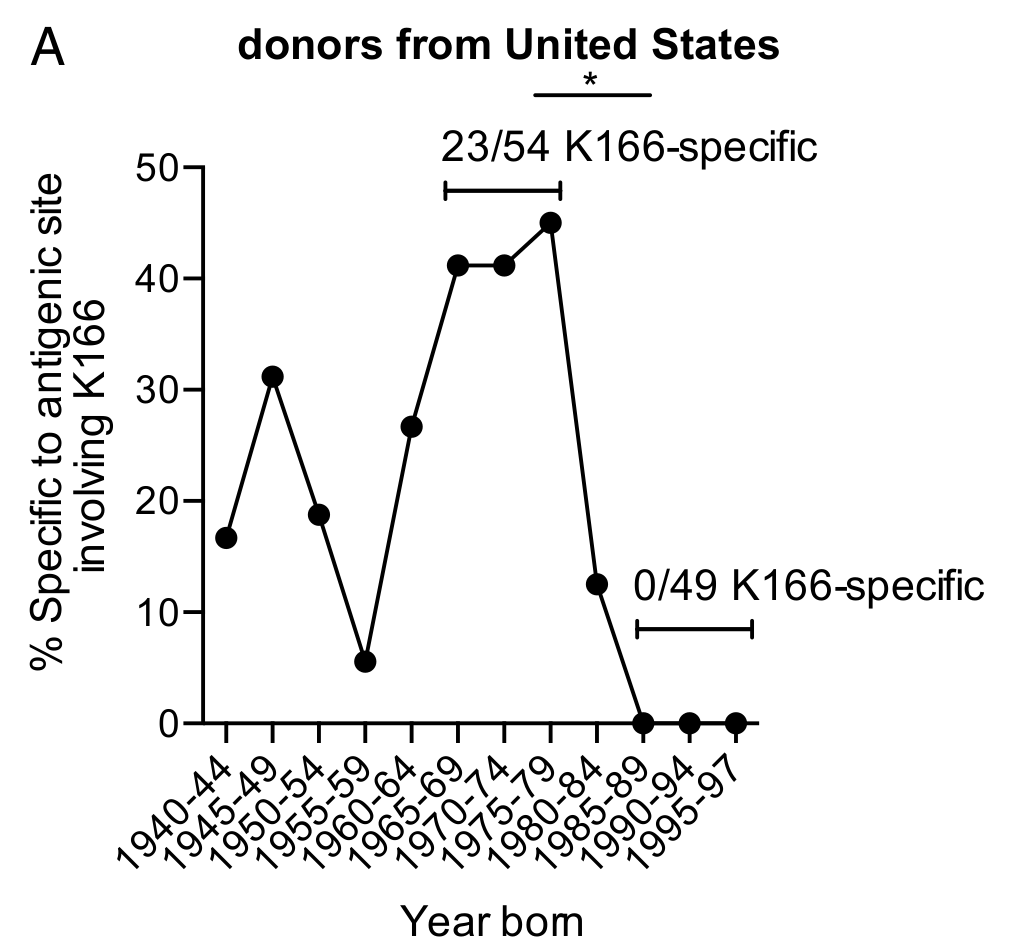 Figure from Linderman et al
Figure from Linderman et al
Human seasonal influenza viruses

Shortcomings of ferret HI data
 Figure from Linderman et al
Figure from Linderman et al
Linderman .... Hensley, PNAS 2014
- People born between 1960 and 1979 target an epitope containing 163K
- Young people target a different epitope -- like ferrets
- The 163K epitope in H1N1pdm is similar to H1N1 viruses circulating 1976-1986
- The vaccine virus was 163K, likely boosted the old response
- After 1986, 163K was shielded by glycosylation
- People born later had no old ABs targeting 166K
Conclusions and Questions
- We need to complement ferrets with systematic human serology
- How do we move away from egg based vaccines?
- How important is the history of exposure to vaccine efficacy?
- What mediates the competition between distant influenza lineages?
- Universal vaccines?
Influenza and Theory acknowledgments




- Boris Shraiman
- Colin Russell
- Trevor Bedford
- Oskar Hallatschek
- All the NICs and WHO CCs that provide influenza sequence data
- The WHO CCs in London and Atlanta for providing titer data



nextflu.org





- Trevor Bedford
- Colin Megill
- Pavel Sagulenko
- Sidney Bell
- James Hadfield



