Recent progress in understanding evolutionary dynamics
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/201710_Solvay.html
 Ernst Haeckel, 1879
Ernst Haeckel, 1879
From qualitative to quantitative
- What are the relevant parameters?
- How does adaptation and diversity depend on parameters?
- How repeatable is evolution?
- How predictable is evolution?
- How gradual is evolution?
Population genetics & evolutionary dynamics
evolutionary processes ↔ trees ↔ genetic diversity
Concepts and typical assumptions -- 20th century
Assumptions:
- The world doesn't change!
- Mutations happen one at a time!
- Typical events dominate!
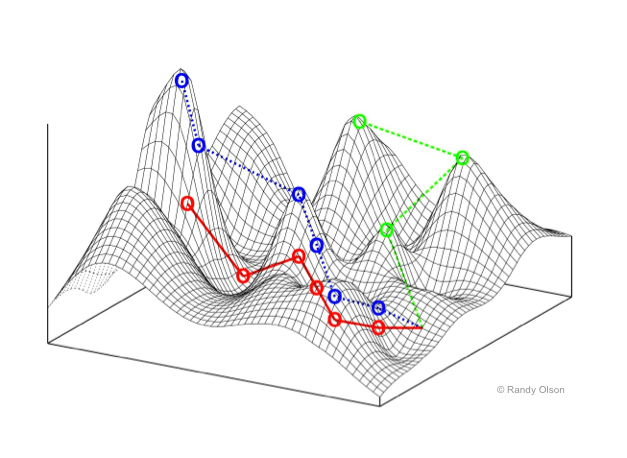
Fitness landscapes
Diffusion theory (Kimura)
$\frac{\partial P(x,t)}{\partial t} = \frac{\partial}{\partial x}\left[-sx(1-x)+\frac{\partial}{\partial x}\frac{x(1-x)}{N}\right]P(x,t)$- Stochastic dynamics of a single locus
- Difficult to generalize to many loci
Coalescence theory (Kingman)
- Backwards in time model
- Predictions for diversity at many loci
- Difficult to generalize to non-neutral variation
Development of sequencing technologies
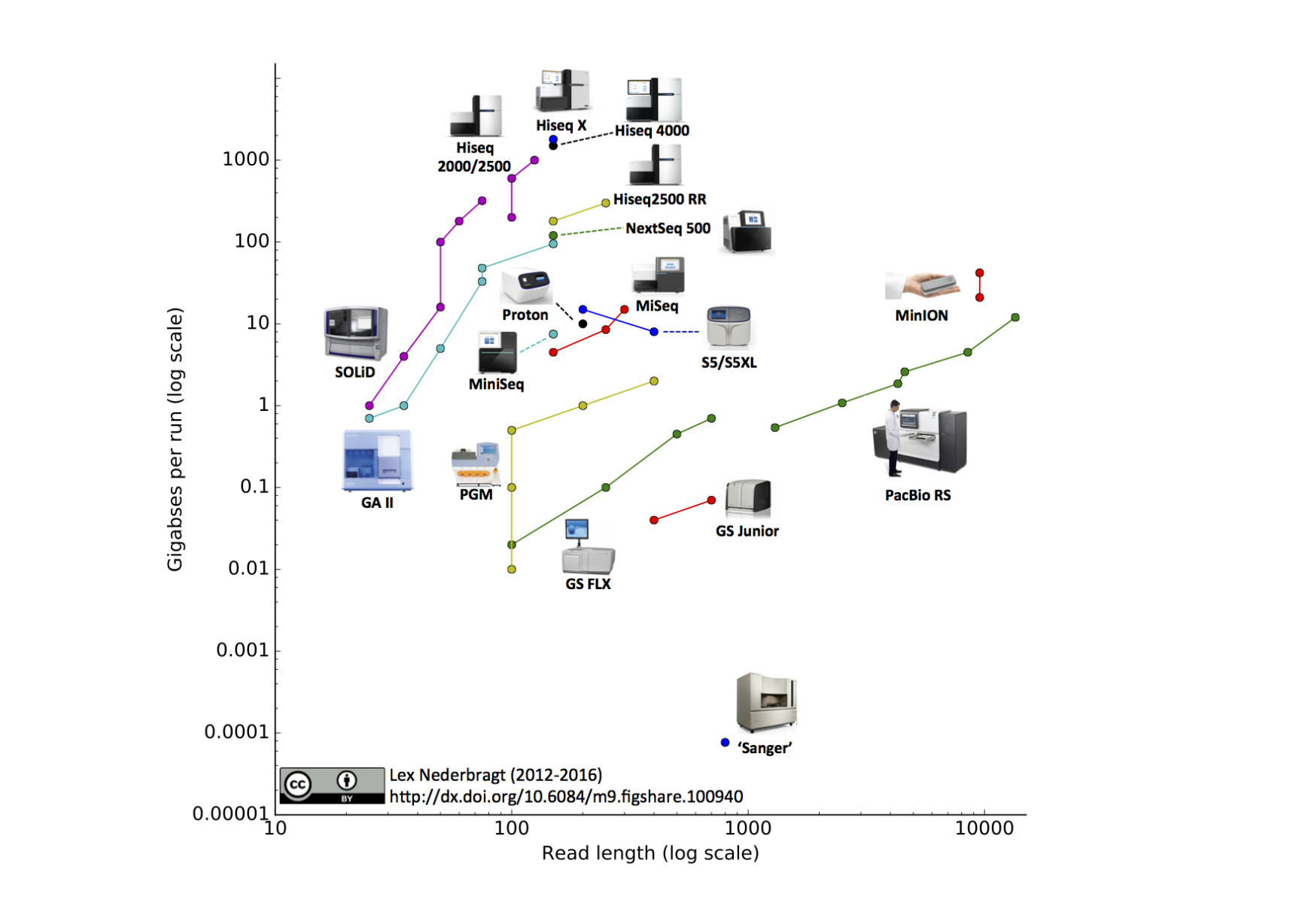
We can now sequence...
- thousands of bacterial isolates
- thousands of single cells
- populations of viruses, bacteria or flies
- diverse ecosystems

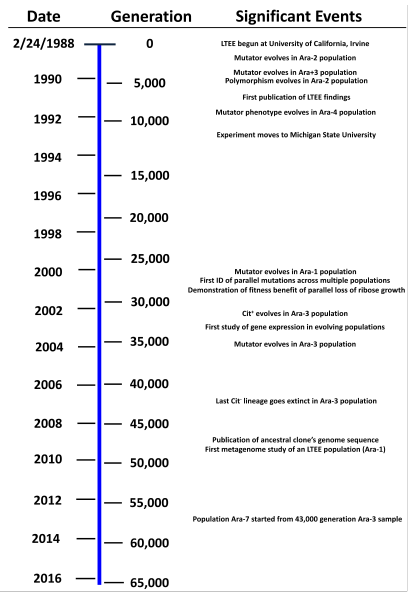
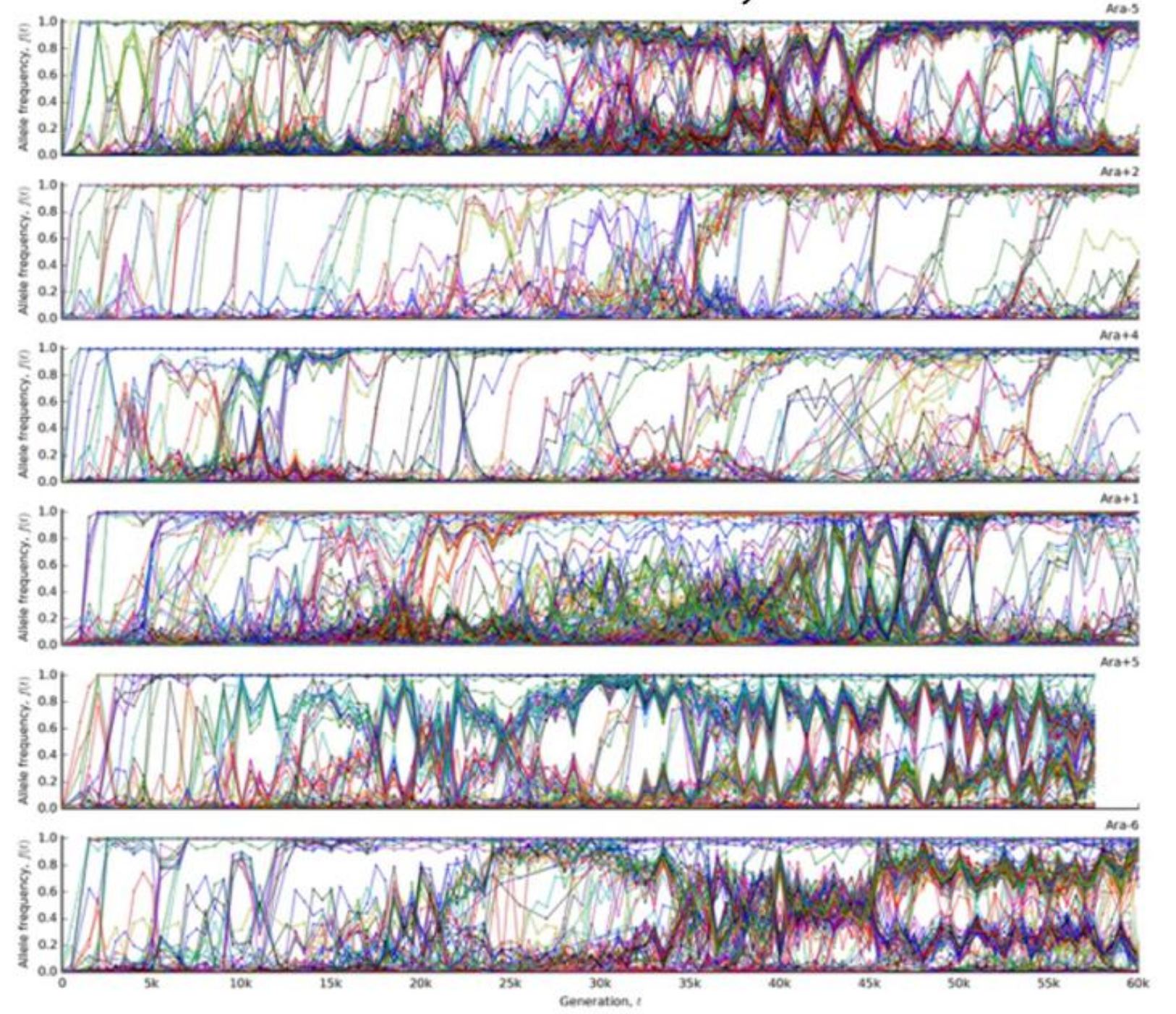
Experimental evolution -- Lenski experiment



Influenza A/H3N2

- Influenza viruses evolve to avoid human immunity
- Vaccines need frequent updates

Rapidly evolving RNA viruses -- HIV
- Many adaptive mutations → mutation supply not limiting
- Many competing clones → complex dynamics
- Environments are constantly changing
Clonal interference and traveling waves
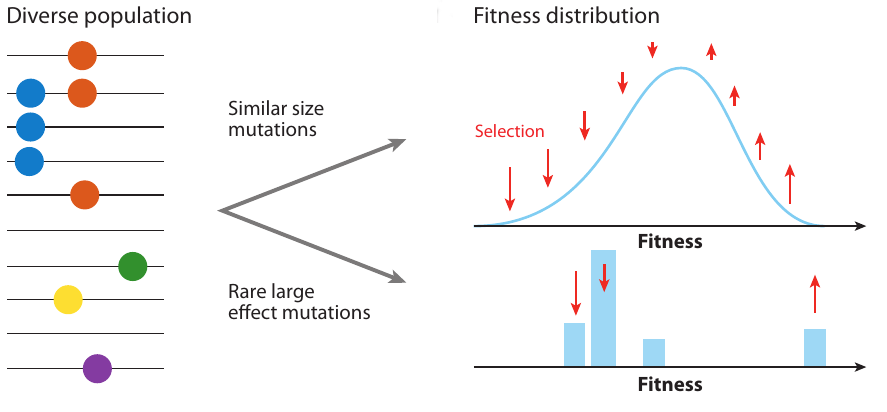
Traveling wave models of adaptation
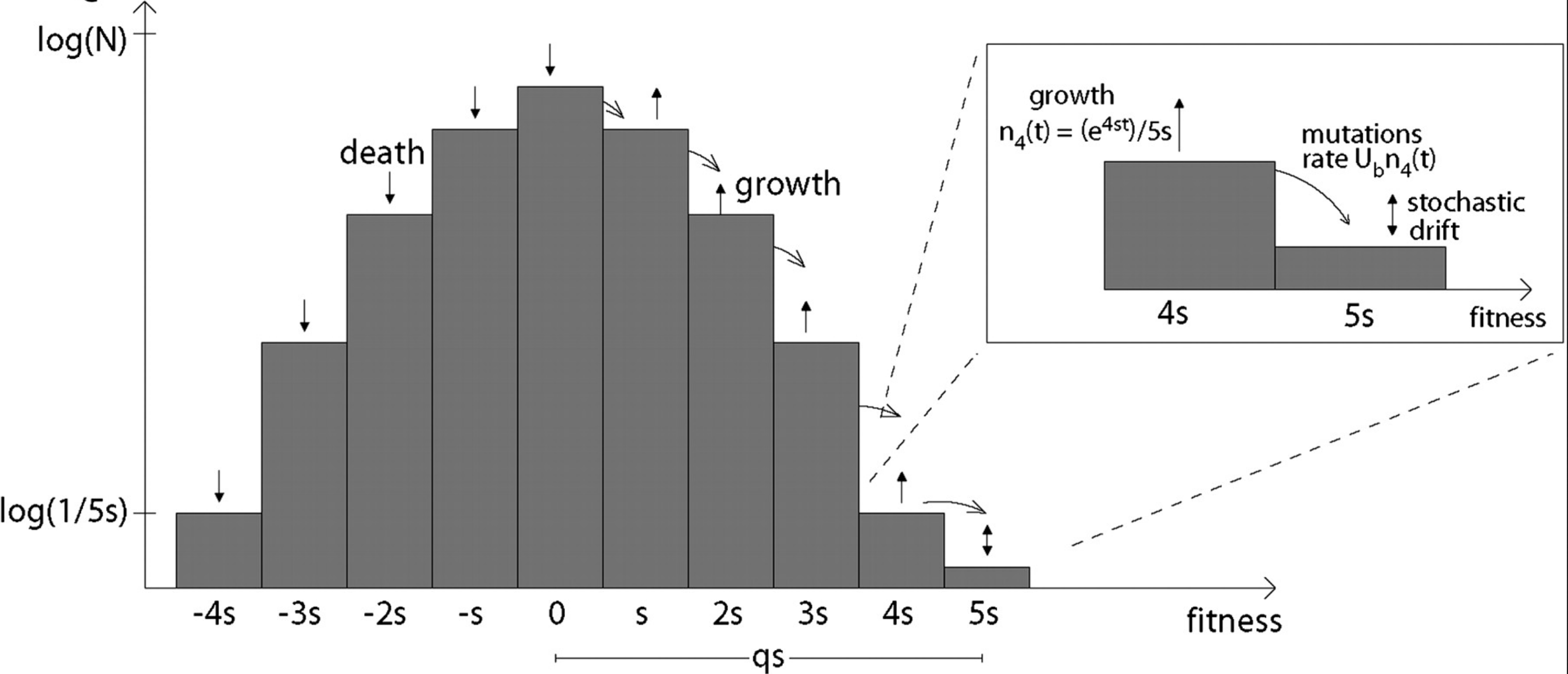
- Speed of adaptation is logarithmic in population size
- Environment (fitness landscape), not mutation supply, determines adaptation
- Different models have universal emerging properties
- Dispersal matters: well mixed populations different from dynamics in one or two dimensions

Bolthausen-Sznitman Coalescent
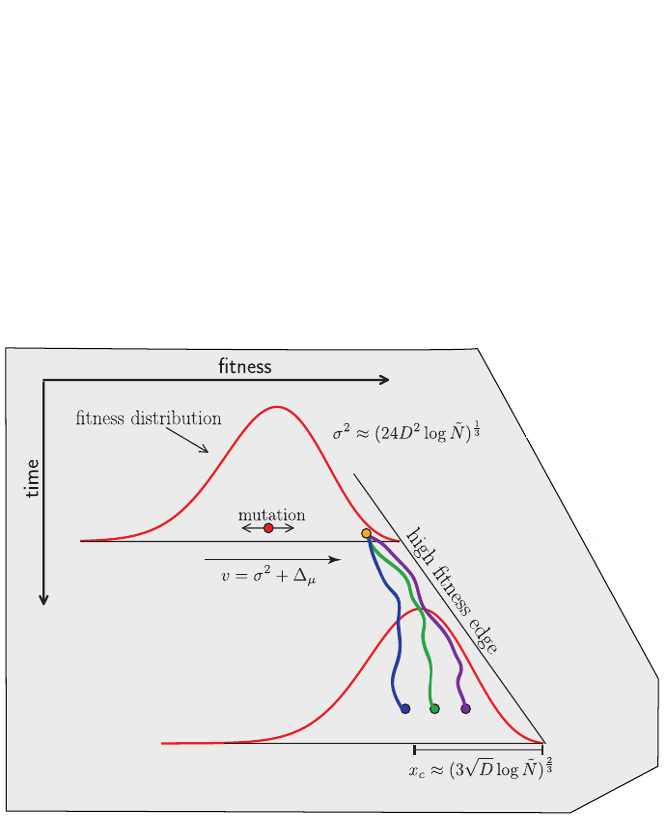
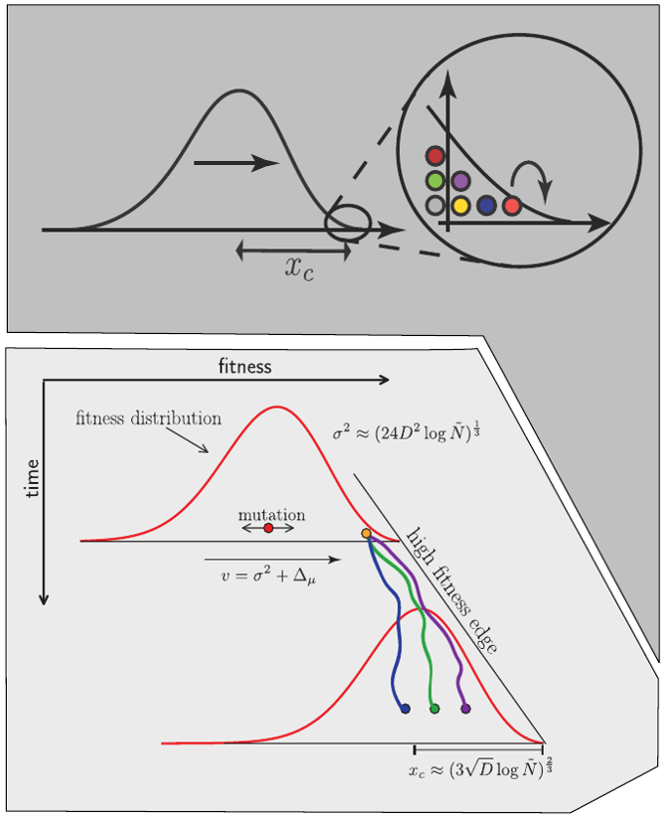
$T_c \sim \log N \quad\mathrm{instead\, of}\quad \sim N$
$\mathrm{rare\,jackpot\,events\,}P(n) \sim n^{-2}$
Brunet and Derrida, PRE, 2007; RN, Hallatschek, PNAS, 2013; Desai, Walczak, Fisher, Genetics, 2013
$\mathrm{rare\,jackpot\,events\,}P(n) \sim n^{-2}$
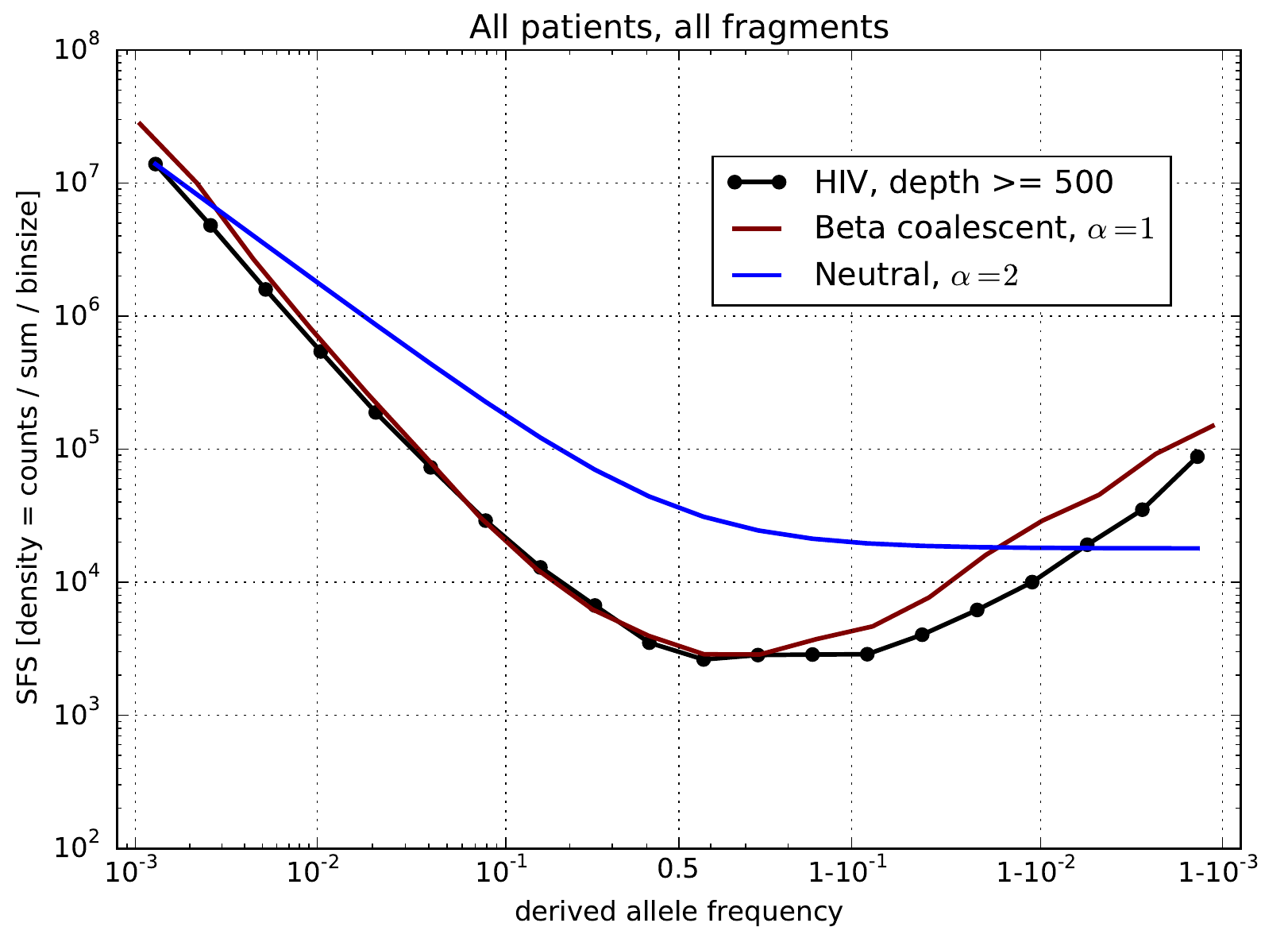
Testable predictions -- site frequency spectra
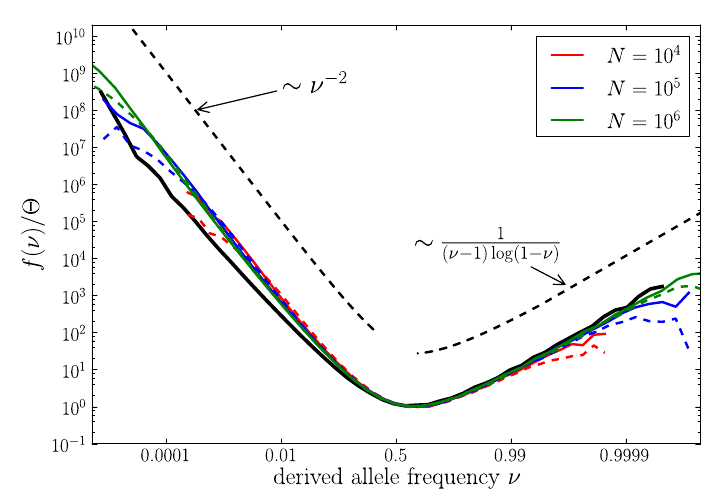

Site frequency spectra observed in HIV populations
Repeatability -- selecting 100 E. coli lines to live at 42C by Tenaillon et al
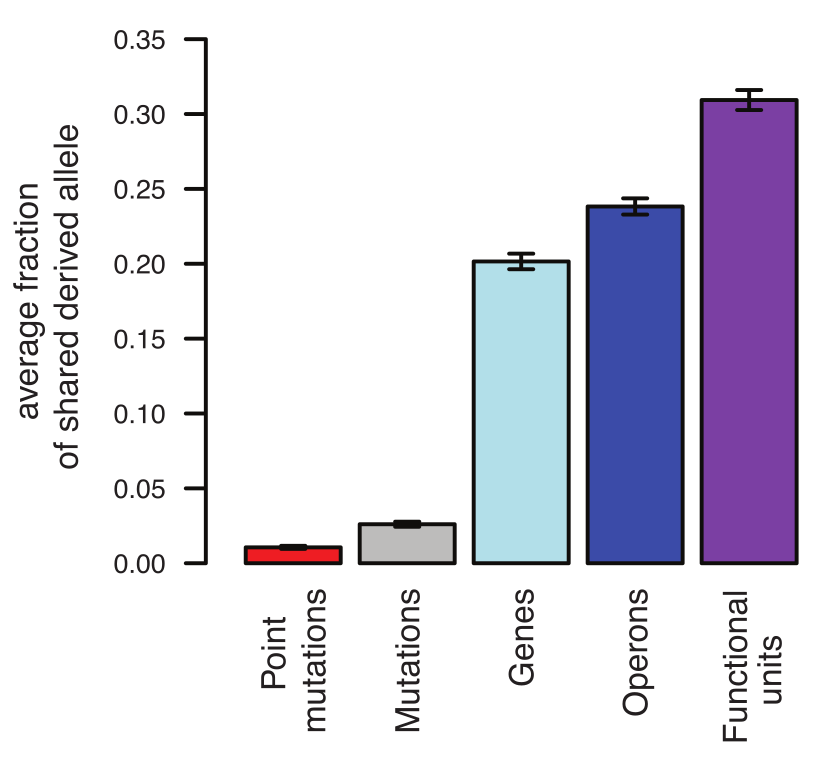 Extensive parallelism at the level of genes and pathways
Extensive parallelism at the level of genes and pathways
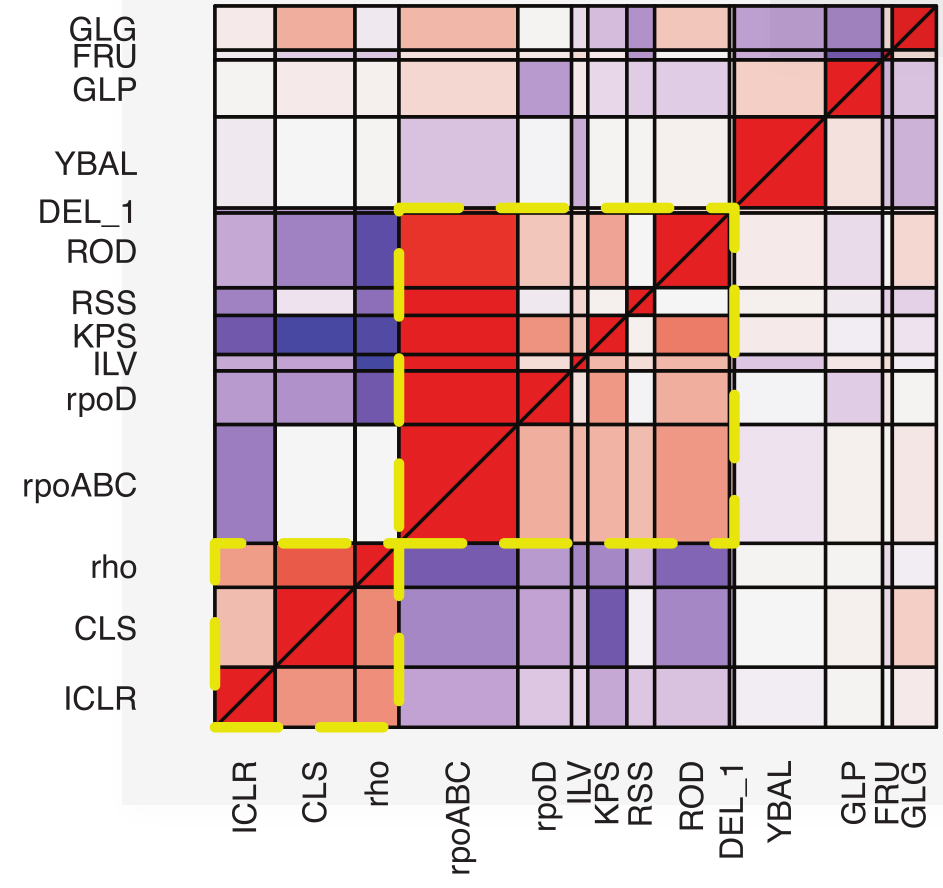 Alternative adaptive path to heat adaptation
Alternative adaptive path to heat adaptation
- Drug resistance: repeated evolution of the same mutations is typical
- Less well defined selection targets: repeatability is restricted to genes or pathways
- Selection on quantitative traits: standing diversity determines response
- Little success in a priori prediction of adaptive pathways
Predicting future populations

General idea
- Future populations descend from present day high fitness individuals
- Identify current high fitness individuals
Predicting influenza
- Approach 1: leverage our understanding of trees of adapting populations
(RN, Russell, Shraiman) - Approach 2: exploit historical patterns indicative of recent adaptation
(Luksza and Lässig) - These methods now inform the WHO vaccine strain selection process.
Conclusions, limitations, questions
- Good understanding of adapting populations under directional selection
- Sequencing is easy, structure and function are hard
- Limited ability to predict the nature of adaptations
- Experimental evolution = artificial environments
→ atypical adaptation, dominated by loss of function mutations.
Is this representative? Better/other systems? - Observable pathogen evolution → Coevolution.
Is this a more general setting (or limited RNA to viruses)?
Beyond one pathogen/one host? - Ecology gets in the way → coevolution, niche construction, variable environments
How does adaptation interact with ecology and intra-species partitioning?
Can we ever separate the environment from adaptation? - How does short term adaptation extrapolate to processes on longer time scales?