Evolution of HIV
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/201712_ICTP1.html
 Ernst Haeckel, 1879
Ernst Haeckel, 1879
From qualitative to quantitative
- What are the relevant parameters?
- How does adaptation and diversity depend on parameters?
- How repeatable is evolution?
- How predictable is evolution?
- How gradual is evolution?
Development of sequencing technologies
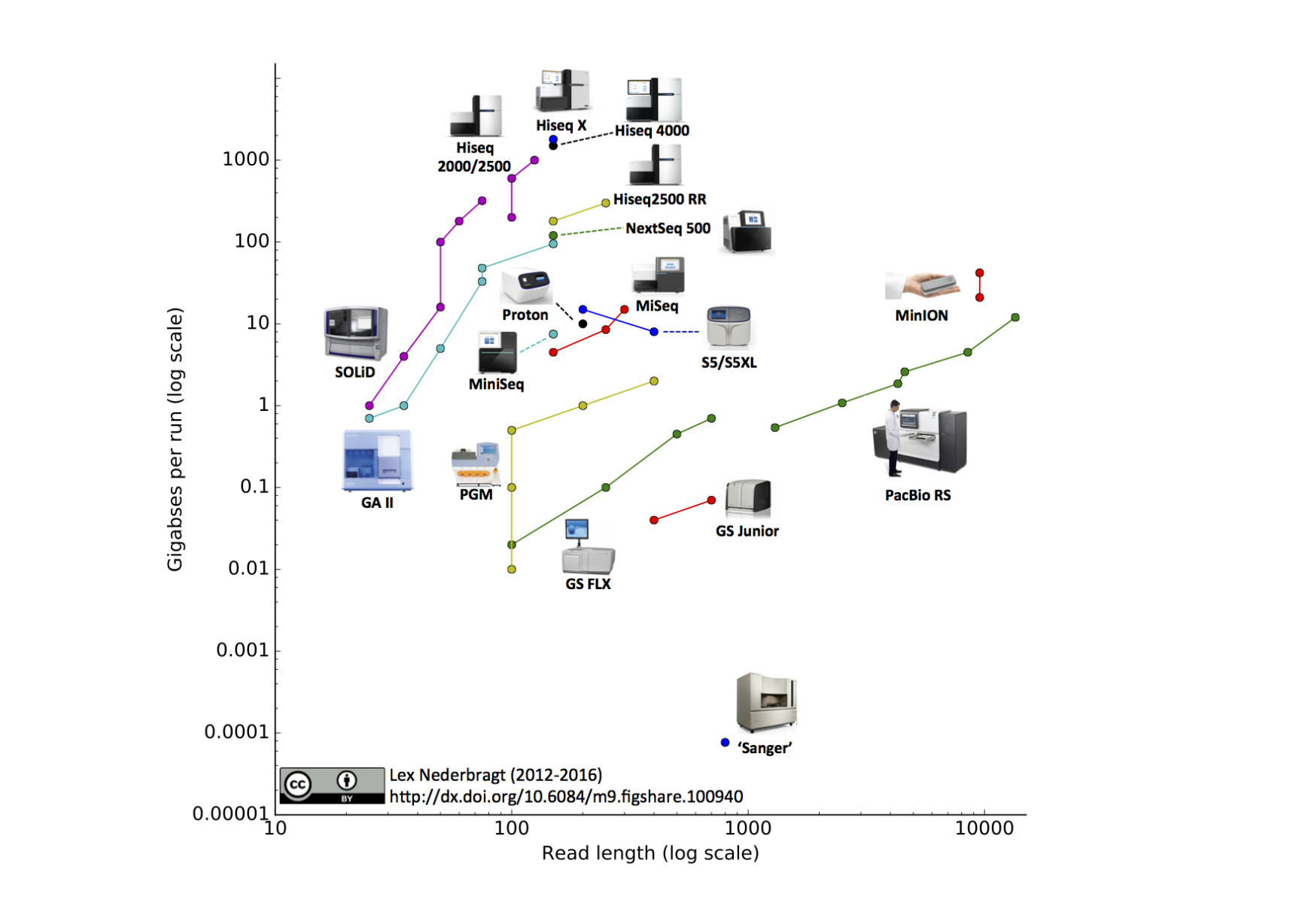
We can now sequence...
- thousands of bacterial isolates
- thousands of single cells
- populations of viruses, bacteria or flies
- diverse ecosystems
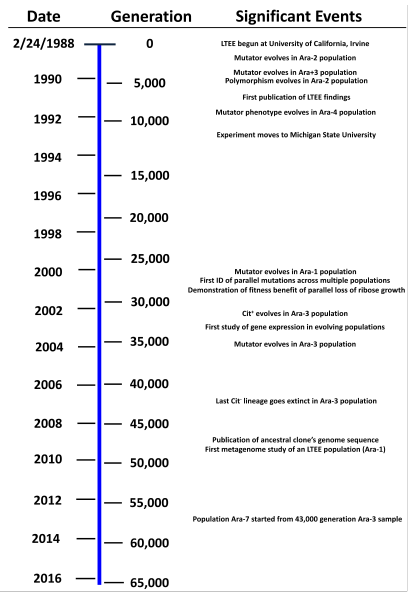
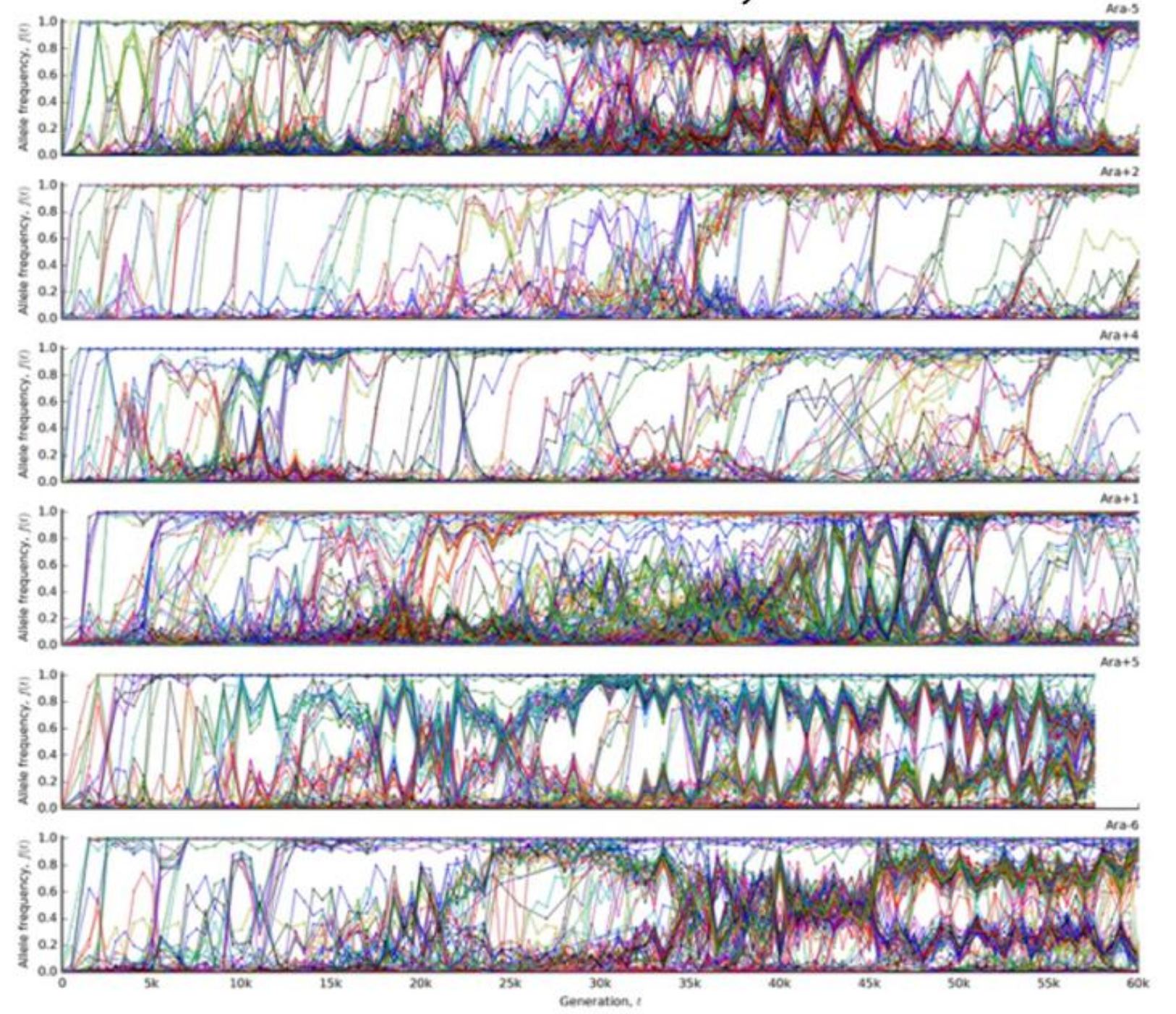
Experimental evolution -- Lenski experiment

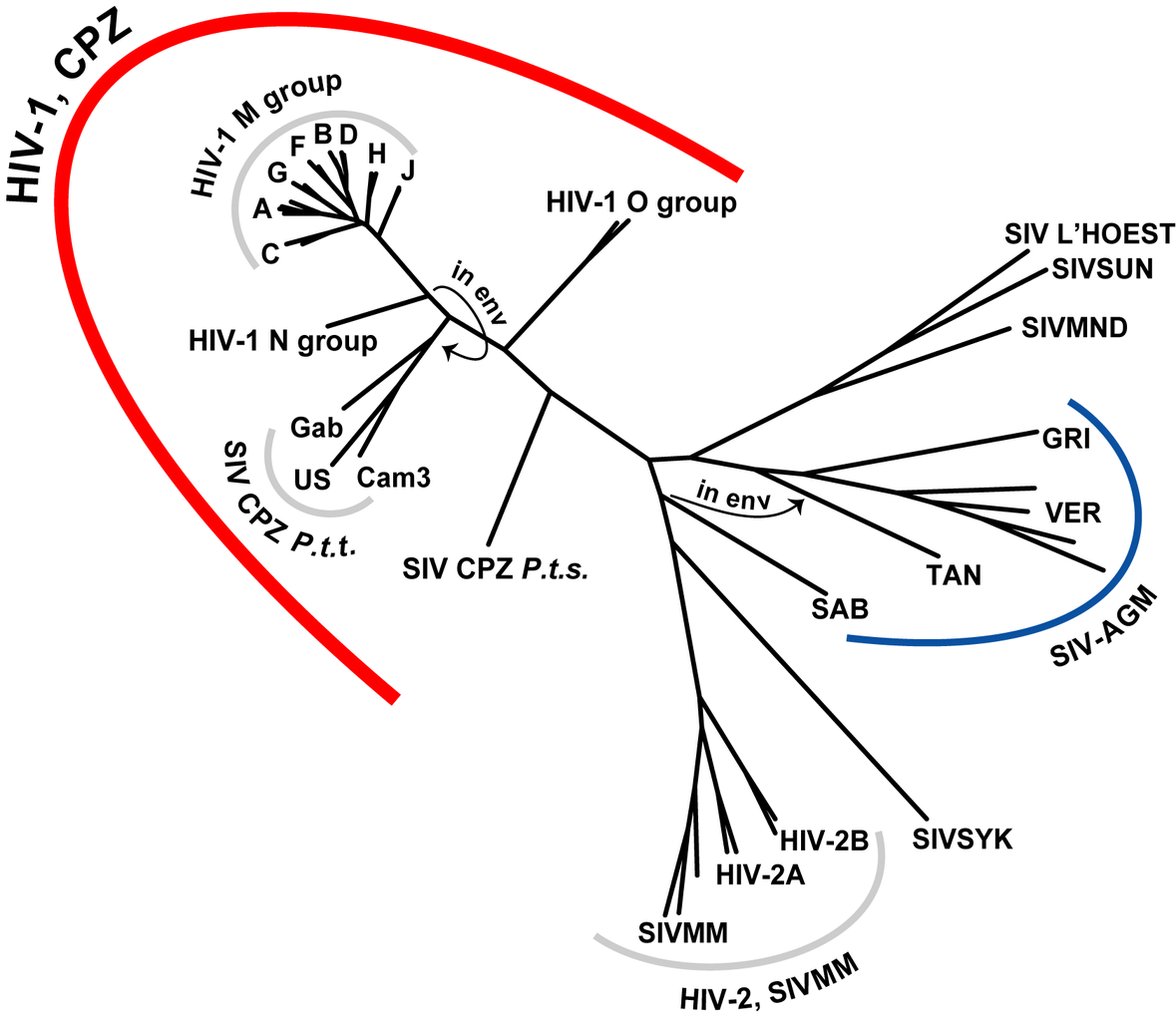
Evolution of HIV
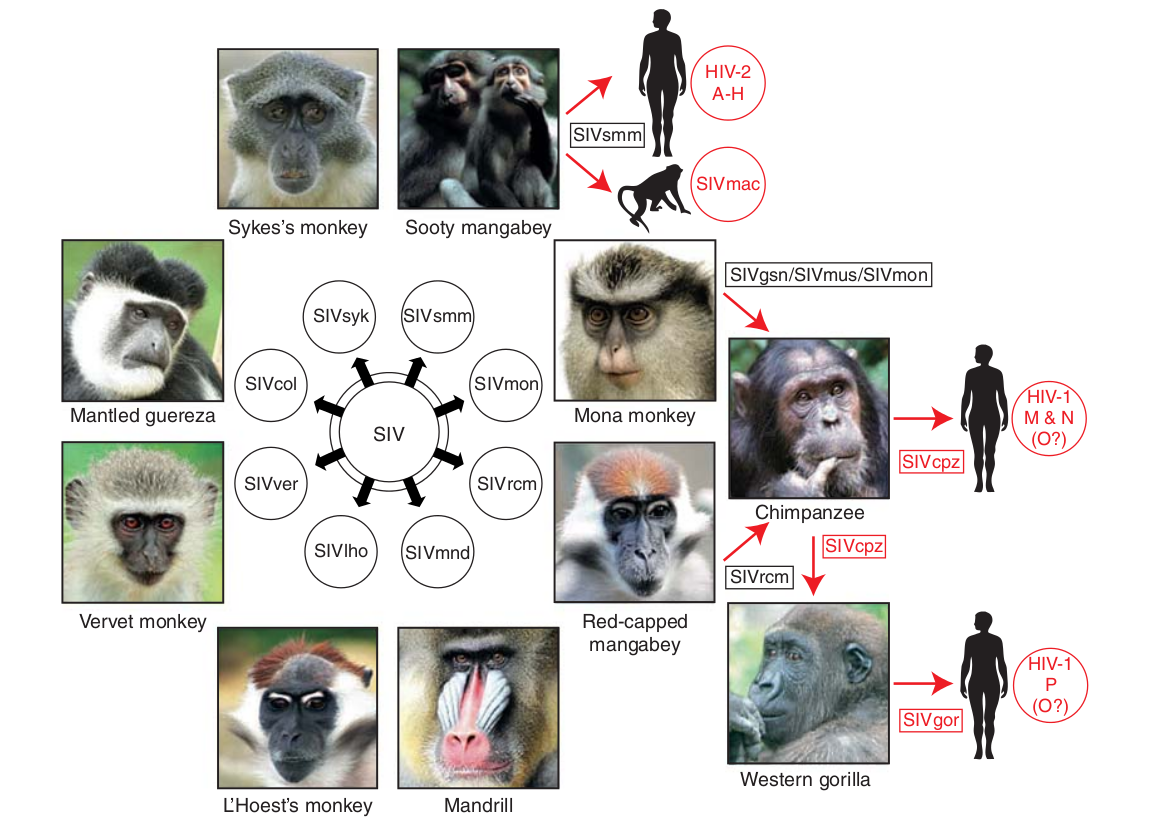
- Chimp → human transmission around 1900 gave rise to HIV-1 group M
- ~100 million infected people since
- subtypes differ at 10-20% of their genome
- HIV-1 evolves ~0.1% per year
Evolution of HIV
HIV infection
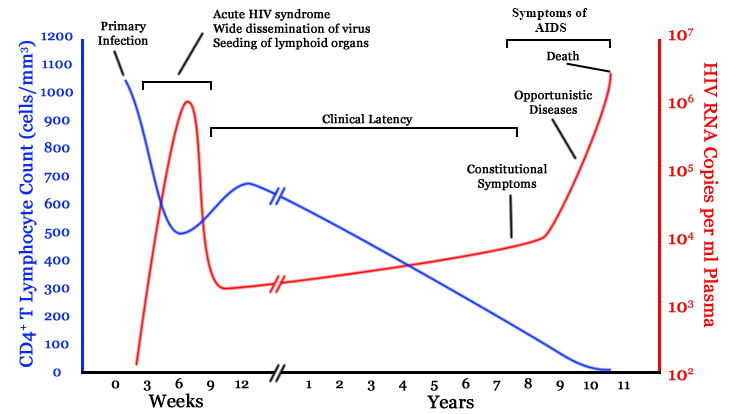
- $10^8$ cells are infected every day
- the virus repeatedly escapes immune recognition
- integrates into T-cells as
latent provirus
HIV-1 evolution within one individual
Immune escape in early HIV infection
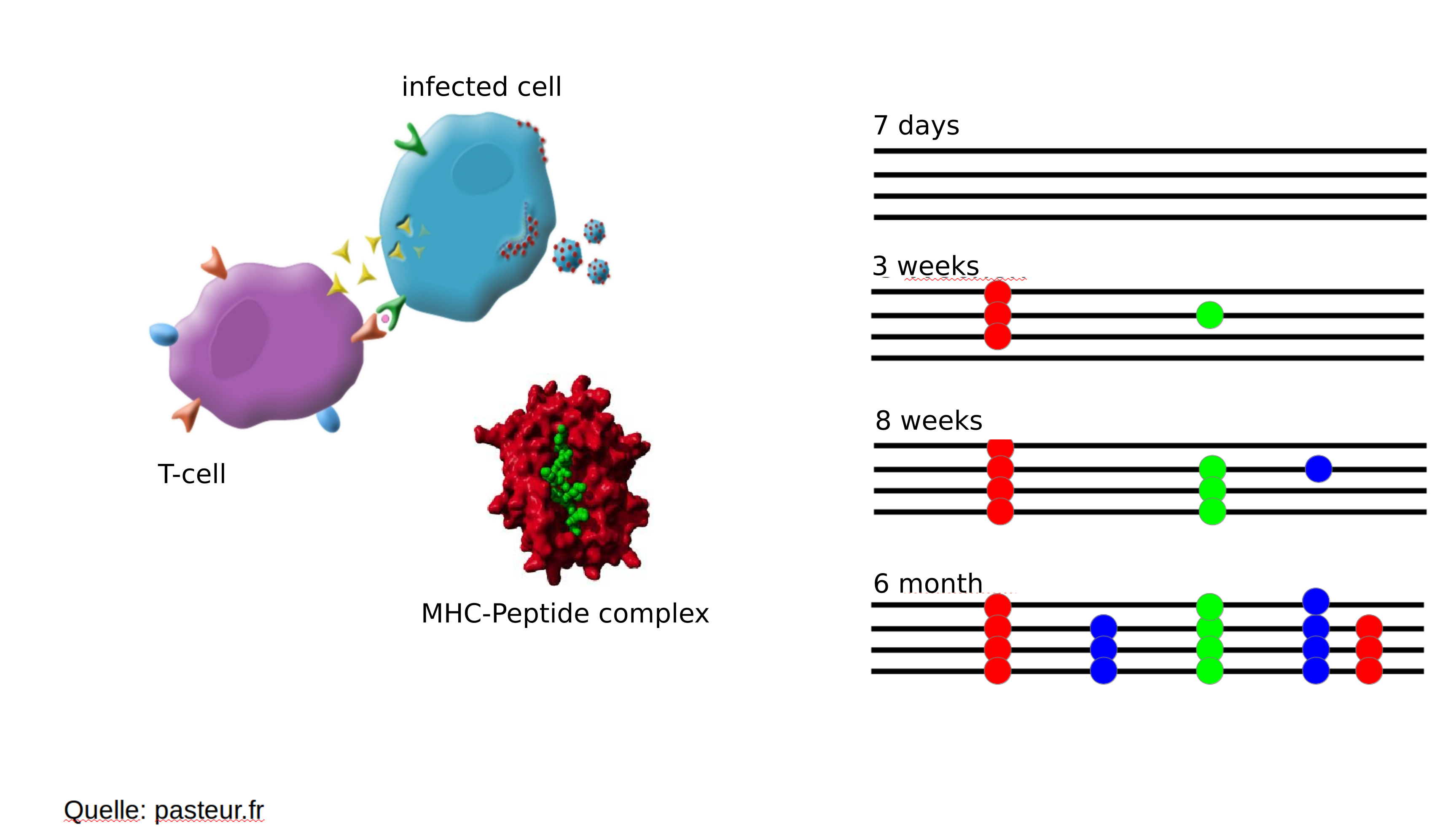
Immune escape in early HIV infection
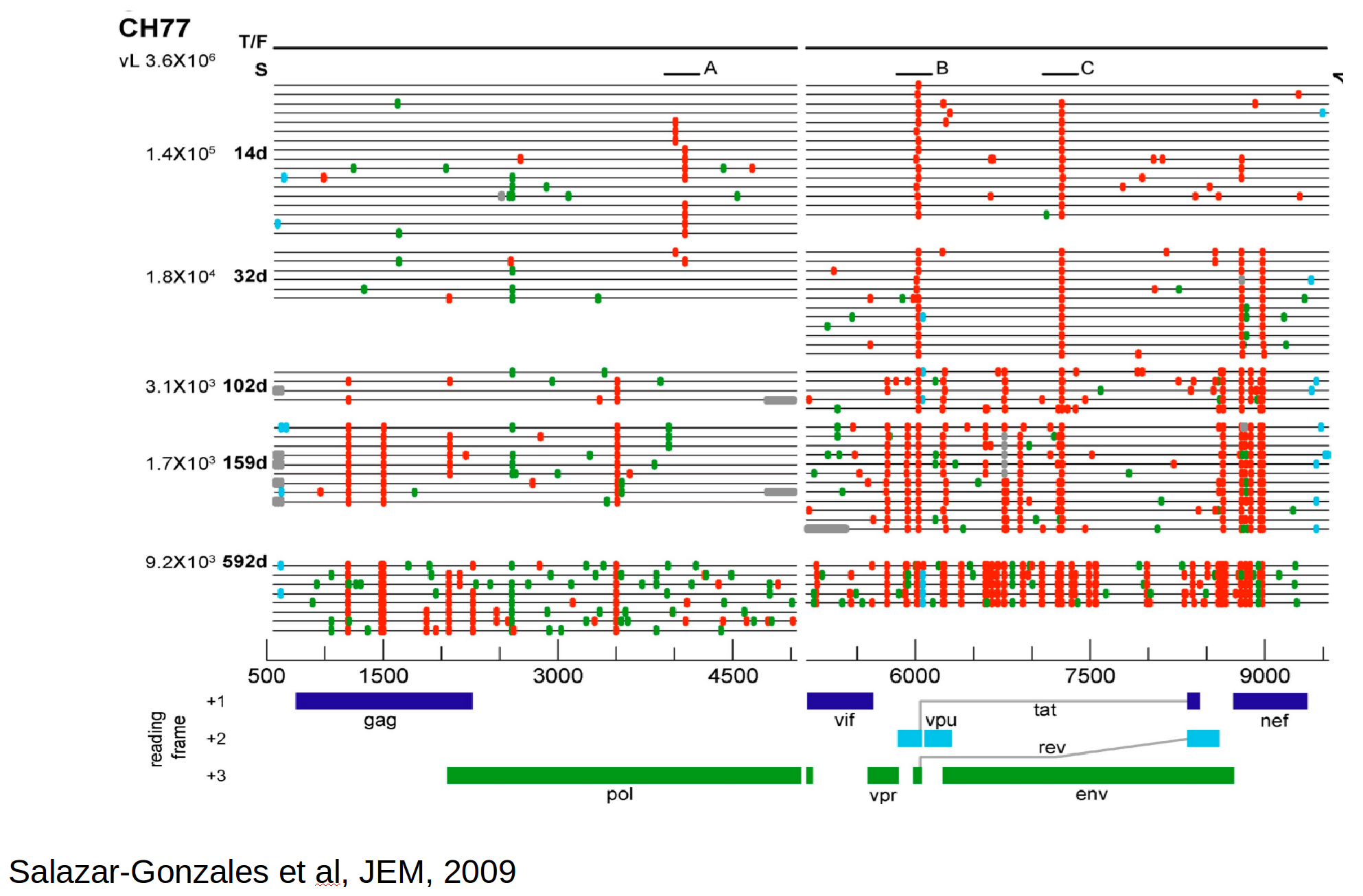
HIV-1 evolution within one individual
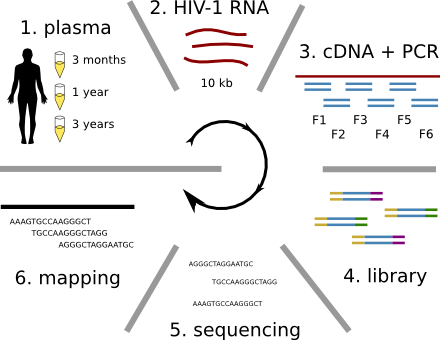

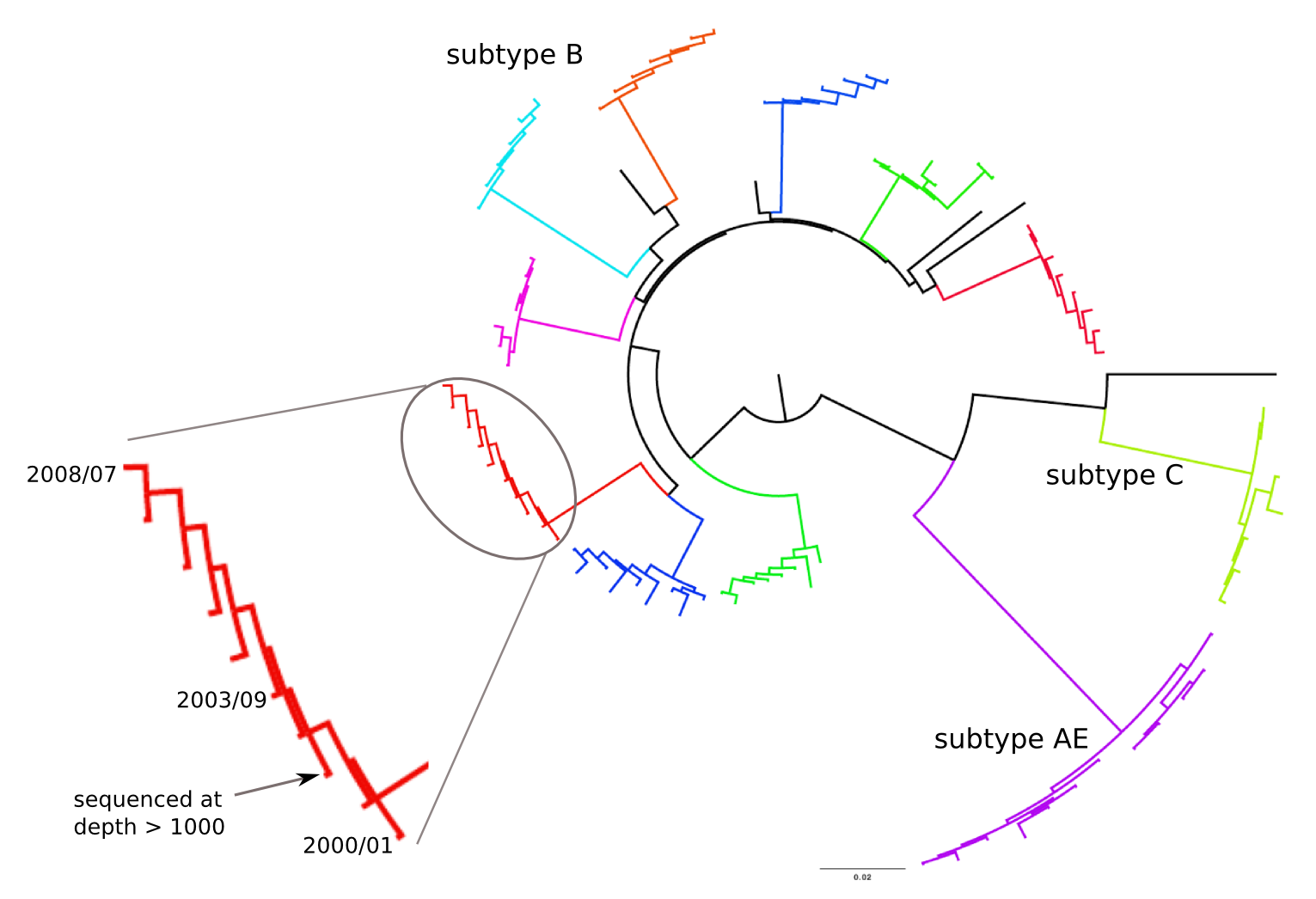
Population sequencing to track all mutations above 1%
- diverge at 0.1-1% per year
- almost whole genome coverage in 10 patients
- full data set at hiv.tuebingen.mpg.de
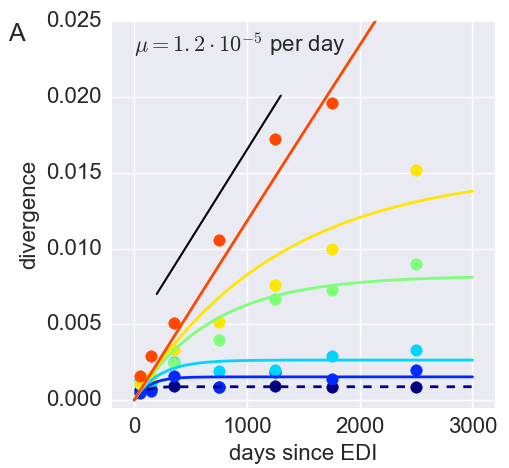
The rate of sequence evolution in HIV
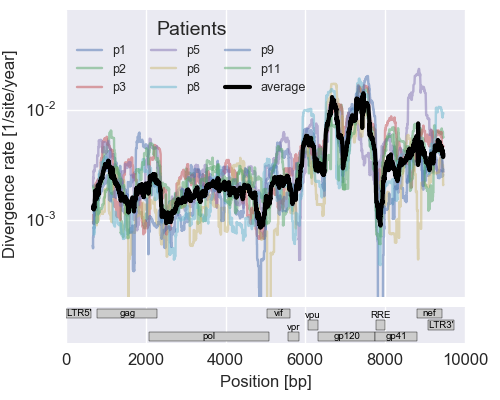
Mutation rate, evolutionary rate, substitution rates
- Mutation rate != evolutionary rate
- Neutral divergence = $\mu t$
- Diversifying selection → faster
- Purifying selection → slower
Evolution in different parts of the genome
- envelope changes fastest, enzymes lowest
- identical rate of synonymous evolution
- diversity saturates where evolution is fast
- synonymous mutations stay at low frequency
Mutation rates and diversity and neutral sites
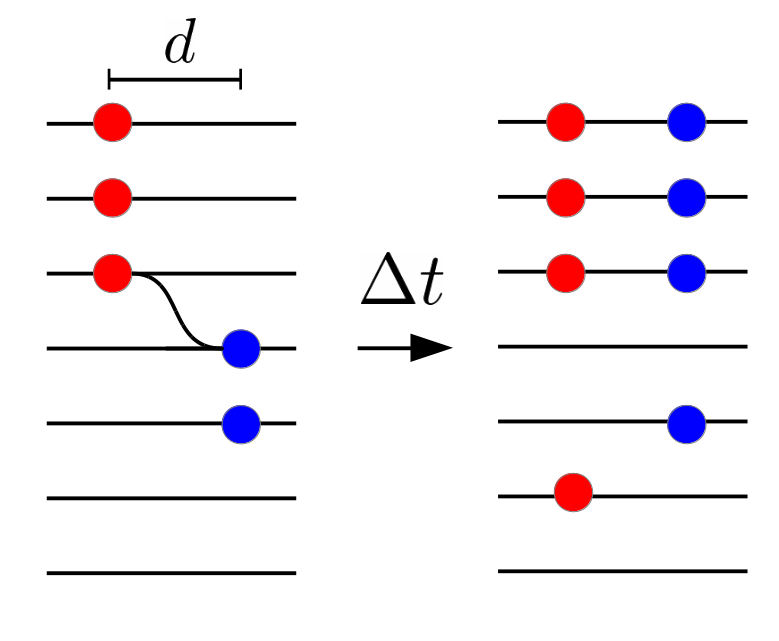
HIV recombination
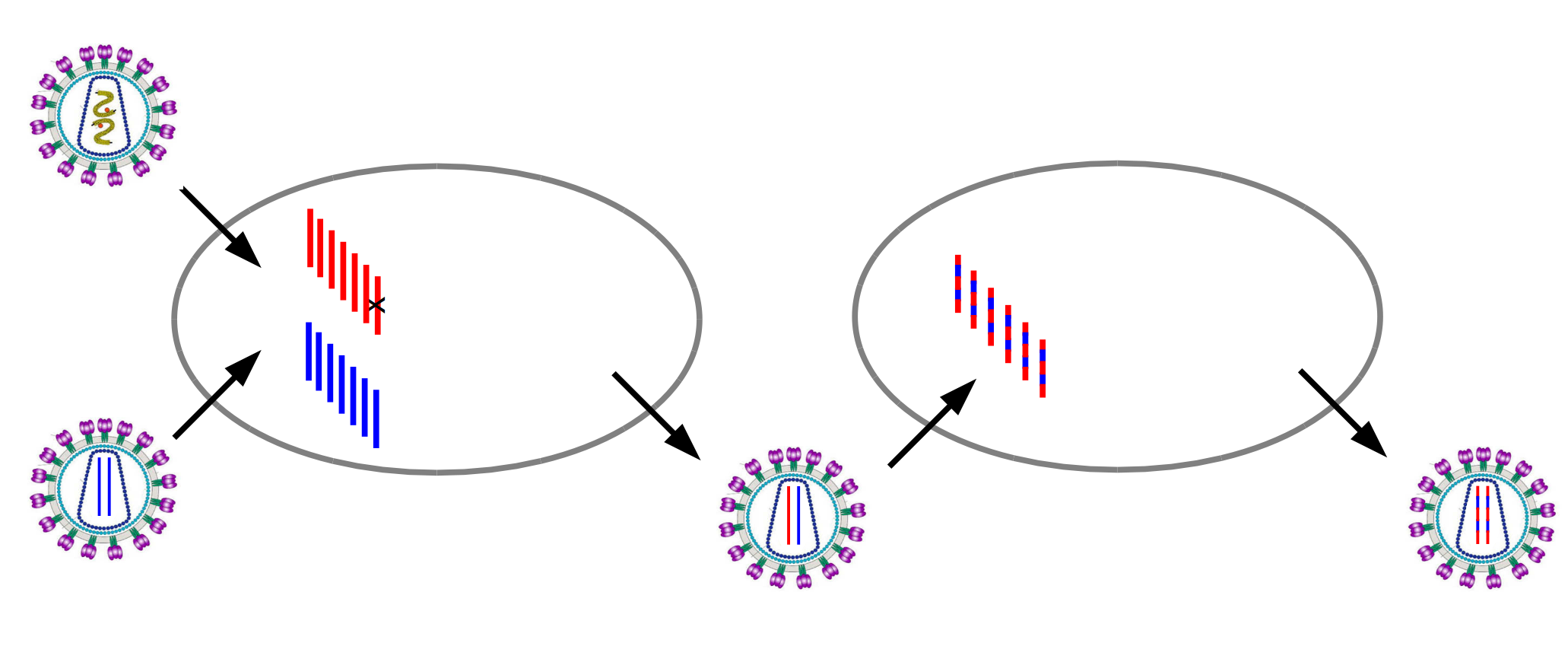
Recombination and linkage
Why recombination?
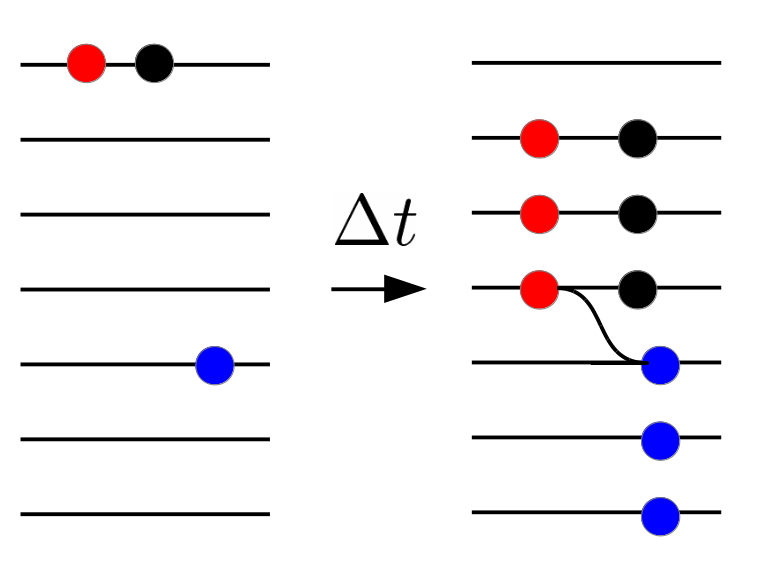
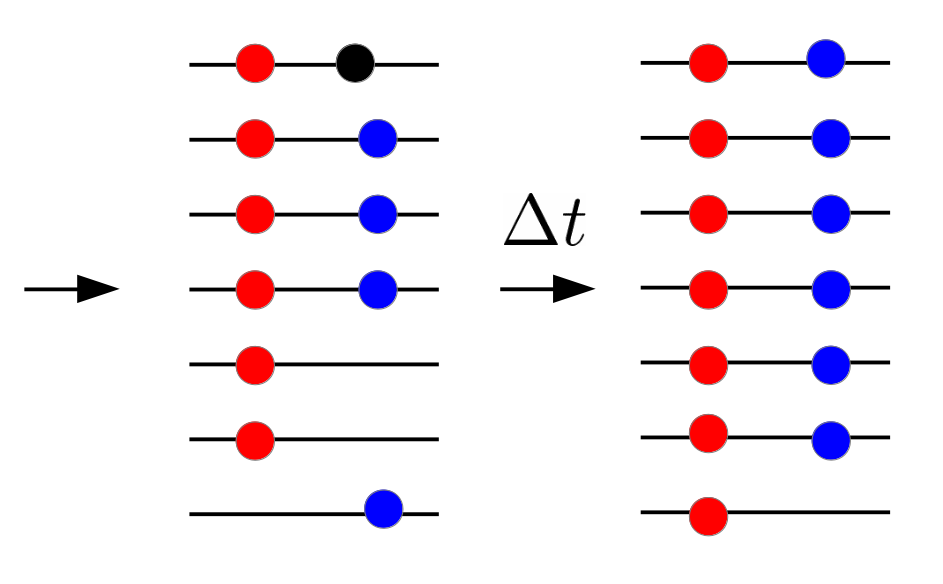
- Red and blue: beneficial mutations
- Black: deleterious mutation
Inference of fitness costs
- mutation away from preferred state at rate $\mu$
- selection against non-preferred state with strength $s$
- variant frequency dynamics: $\frac{d x}{dt} = \mu -s x $
- equilibrium frequency: $\bar{x} = \mu/s $
- fitness cost: $s = \mu/\bar{x}$
Inference of fitness costs
- Frequencies of costly mutations decorrelate fast $\frac{d x}{dt} = \mu -s x $
- $\Rightarrow$ average many samples to obtain accurate estimates
- Assumption: The global consensus is the preferred state
- Only use sites that initially agree with consensus
- Only use sites that don't chance majority nucleotide
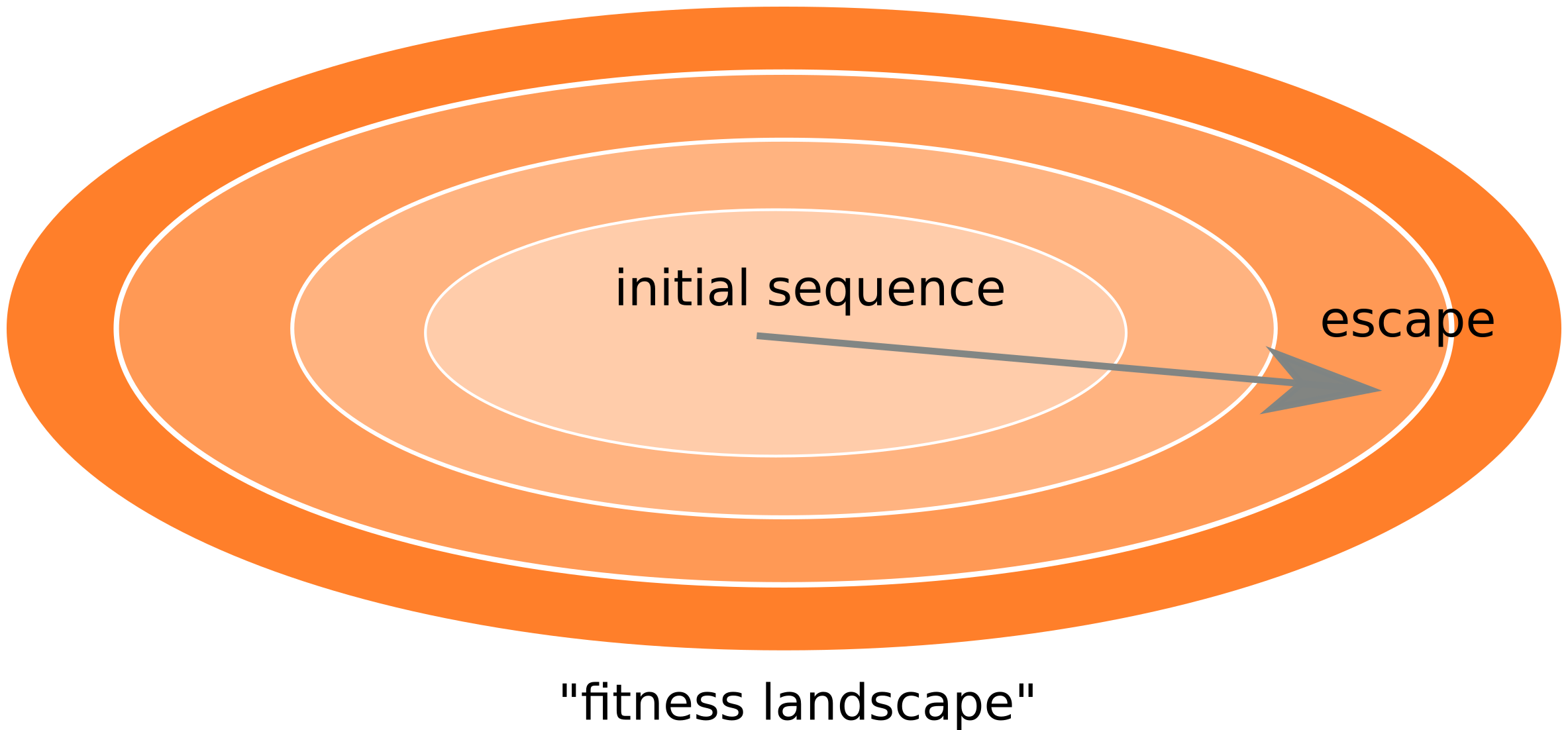
Fitness landscape of HIV-1
Zanini et al, Virus Evolution, 2017Selection on RNA structures and regulatory sites
Zanini et al, Virus Evolution, 2017The distribution of fitness costs
Zanini et al, Virus Evolution, 2017Fitness - diversity correlation
Zanini et al, Virus Evolution, 2017Frequent reversion of previously beneficial mutations
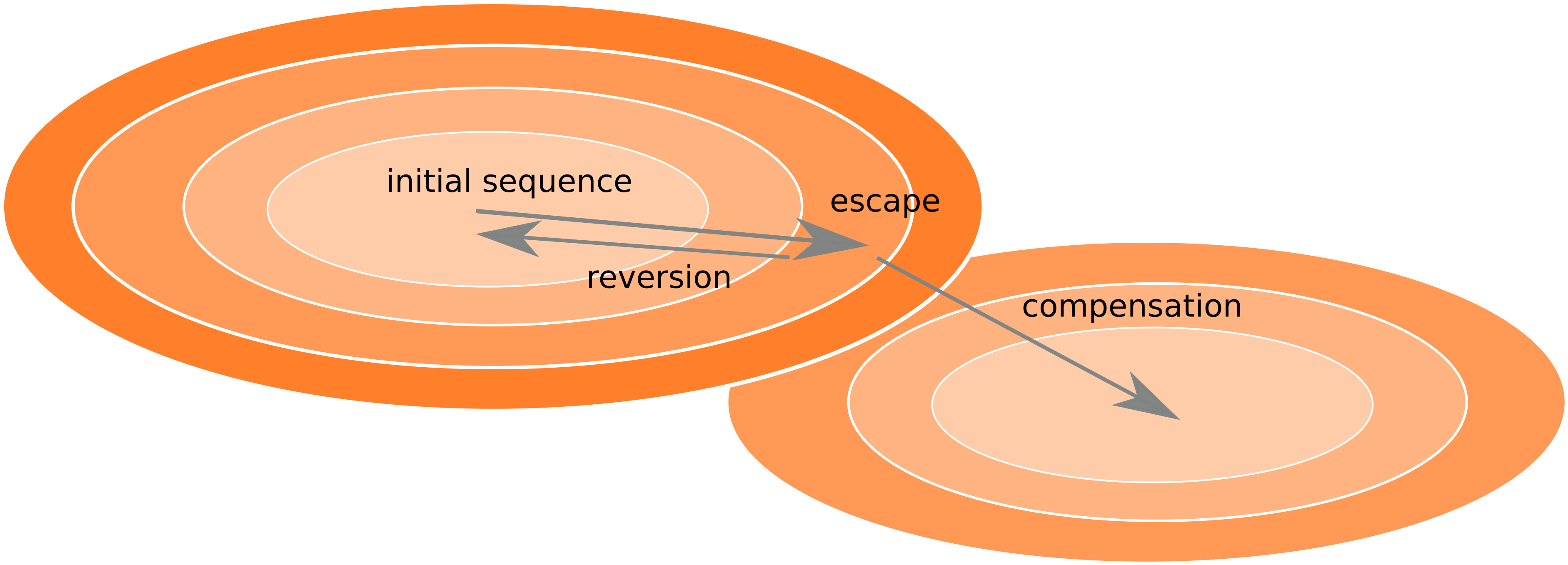
- HIV escapes immune systems
- most mutations are costly
- humans selects for different mutations
- compensation or reversion?

Does HIV evolve during therapy?
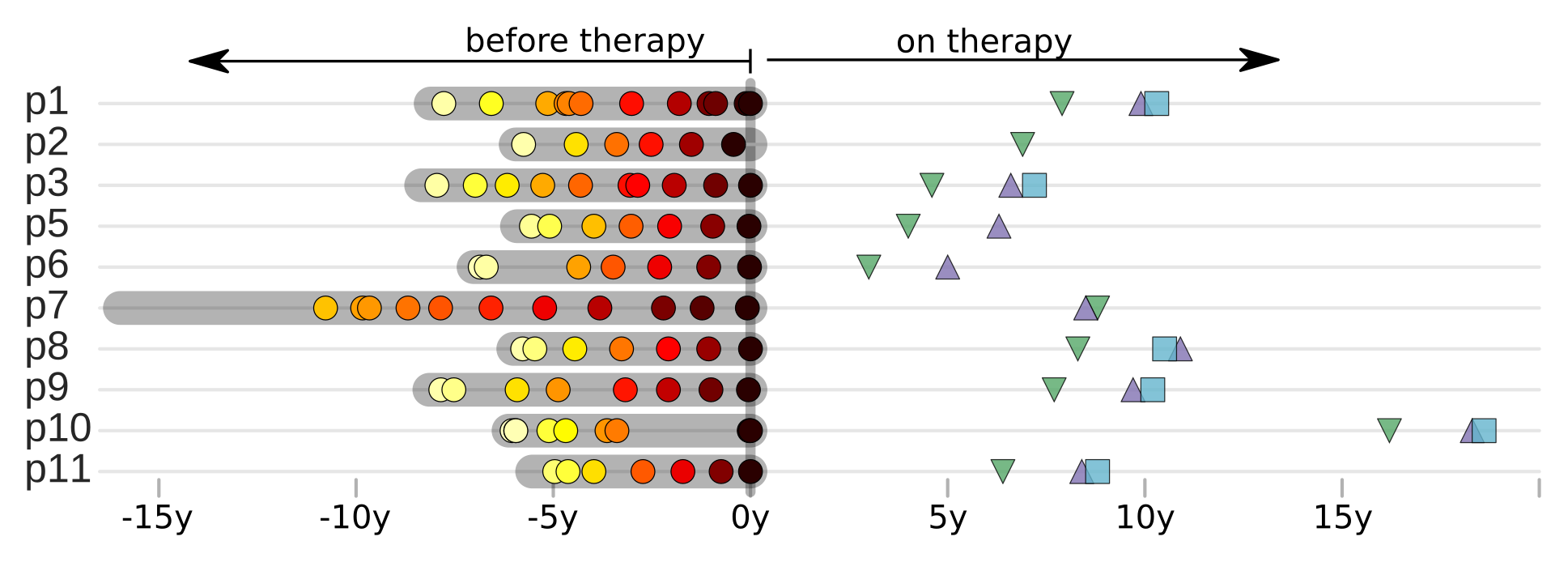 Brodin et al, eLife, 2016
Brodin et al, eLife, 2016
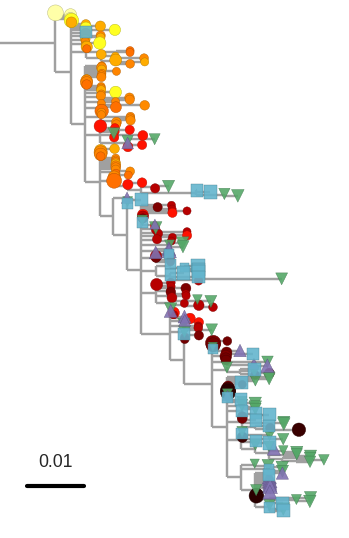
No evidence of ongoing replication
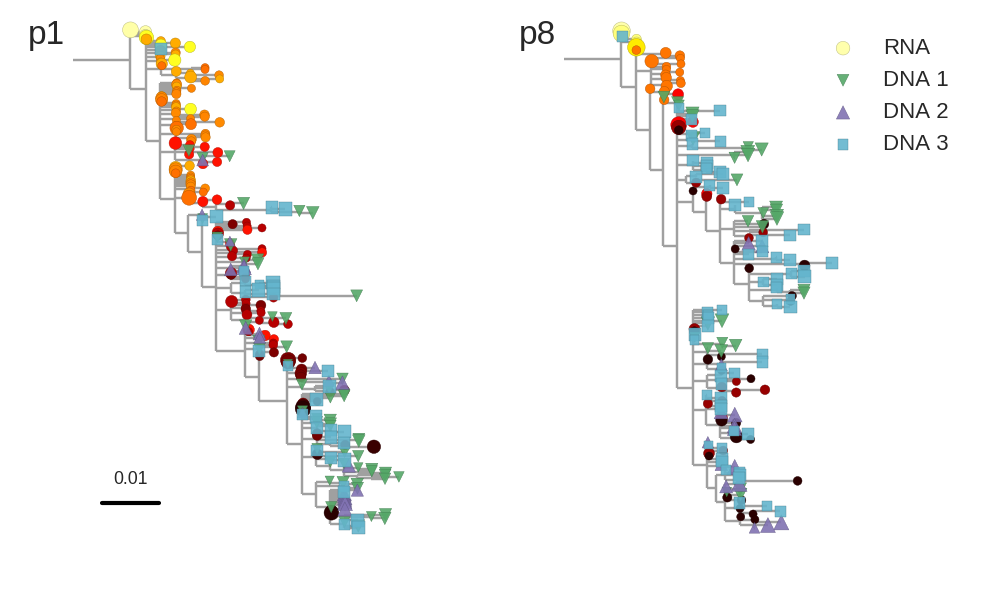
No evidence of ongoing replication
T-cell turnover is fast in untreated infection
- latent HIV → barcode of a T-cell lineage
- all latent integrated virus derives from late infection
- untreated: T-cell lineages are short lived
- on therapy: T-cell clones live decades
Summary
- Deep sequencing allows comprehensive characterization of evolving populations
- RNA viruses evolve rapidly enough to observe dynamics
- Fitness landscape can be estimated explicitly
- Constant struggle between maintaining a functional virus and adaptation to host immunity
- Latently integrated virus is an accurate snapshot of the virus before therapy
- Latent virus can serve as barcode for T-cells.
Questions
- Complex dynamics of many mutations: How do they interact?
- What are appropriate theoretical frameworks?
- Can we quantify the effect of recombination on HIV adaptation?
- How important is epistasis?
- Are there regions in the HIV genome that can't escape the immune system?
HIV acknowledgments







- Fabio Zanini
- Jan Albert
- Johanna Brodin
- Christa Lanz
- Göran Bratt
- Lina Thebo
- Vadim Puller


