Real-time tracking, molecular epidemiology, and data sharing
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/201802_bellagio.html
Evolution of RNA viruses
- Constant struggle to adapt to changing environments and host immunity
- Mutation rates of about 0.00001/site and replication
- Large viral populations explore every single mutation every day
- Most mutations are detrimental, but some persist
- Mutations can act like an approximate clock
Phylogenetic analysis
Phylogenetic analysis
Can we use sequences to track an ongoing outbreak?
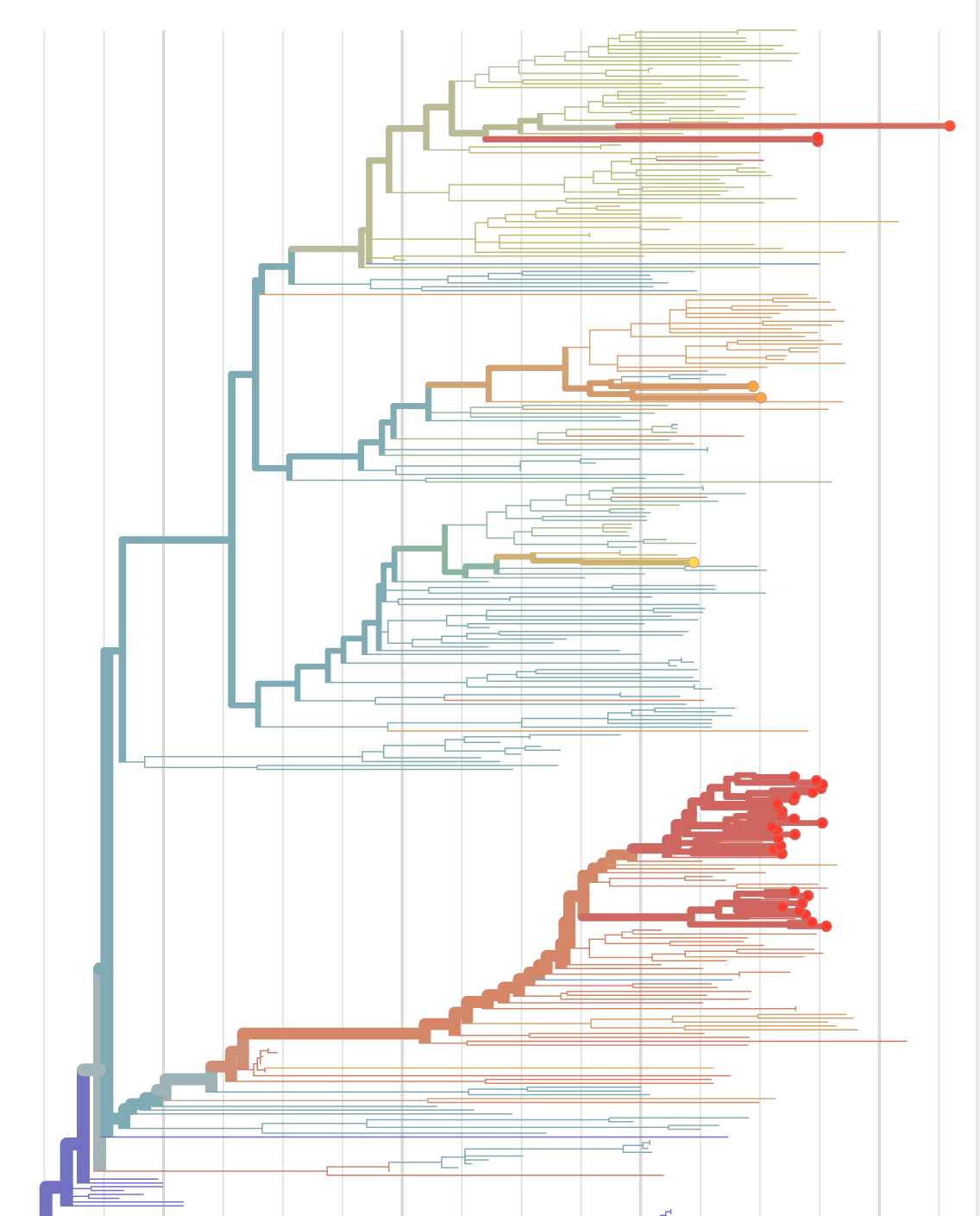
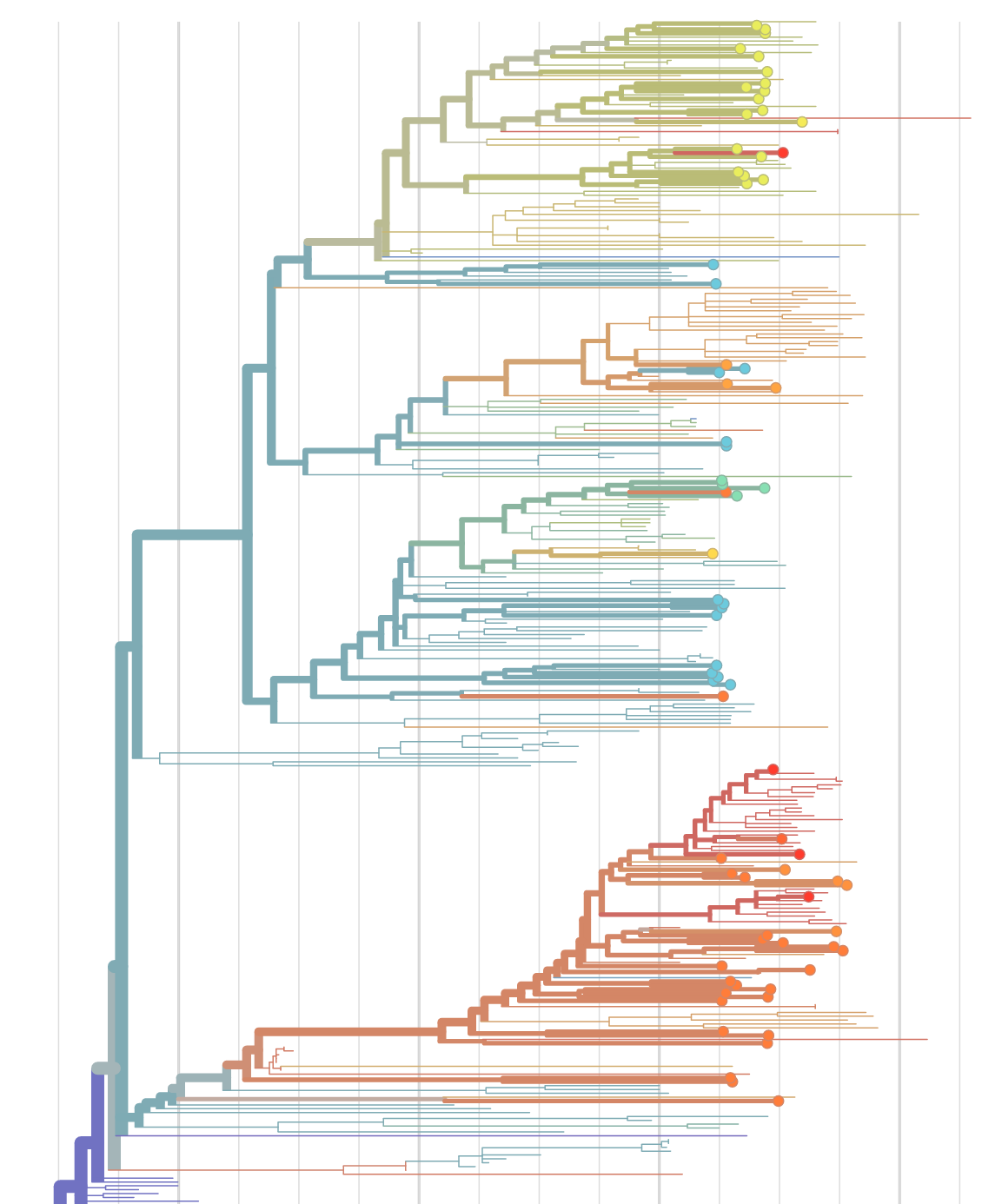
Sequences record the spread of pathogens




The resolution is limited by the number of mutations!
Influenza virus genome - 8 segments
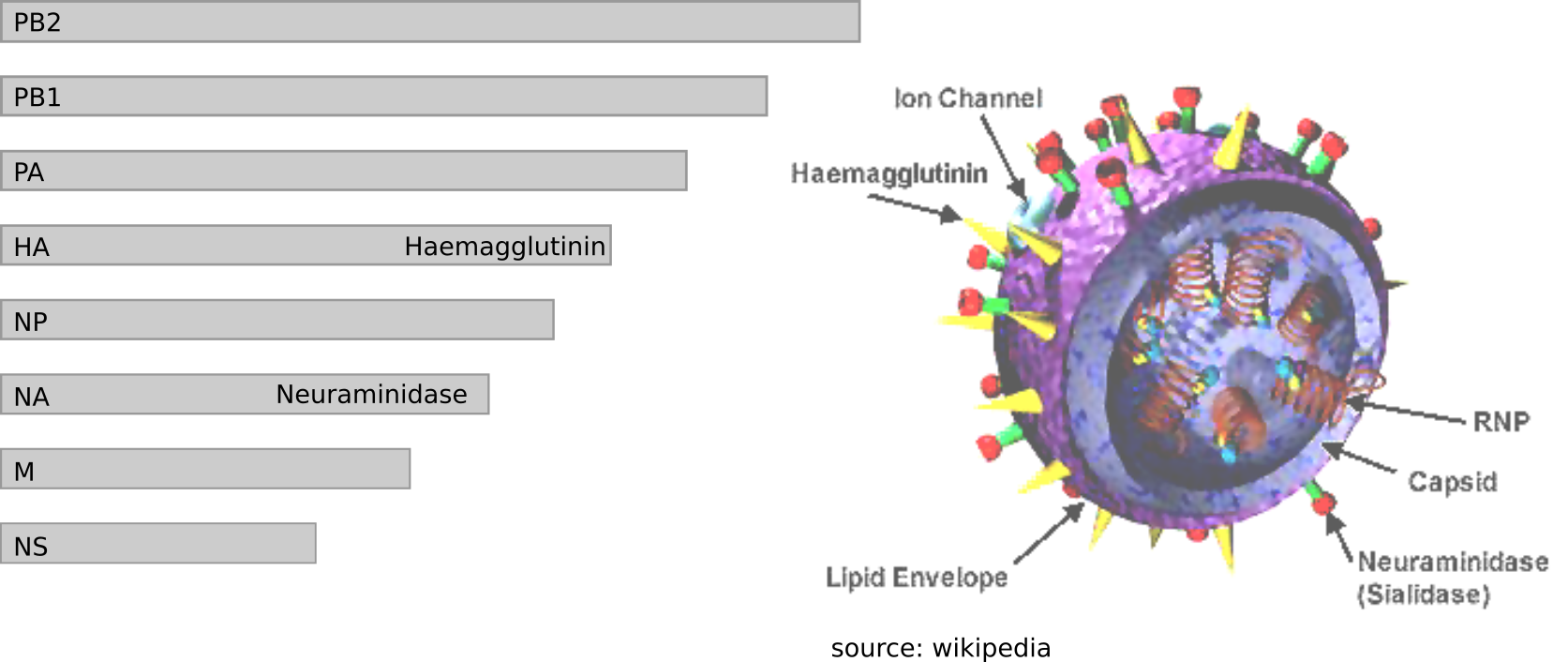
Zika virus genome $\sim 10000$ bases

Ebola virus genome $\sim 20000$ bases

Many RNA viruses pick up one mutation every 2-4 weeks!
Need to sequence the entire genome!
Frequent mutations imply...
- most viruses in an outbreak differ from each other
- data suggest transmission chains
- transmission can be ruled out!
- geographic spread can be reconstructed



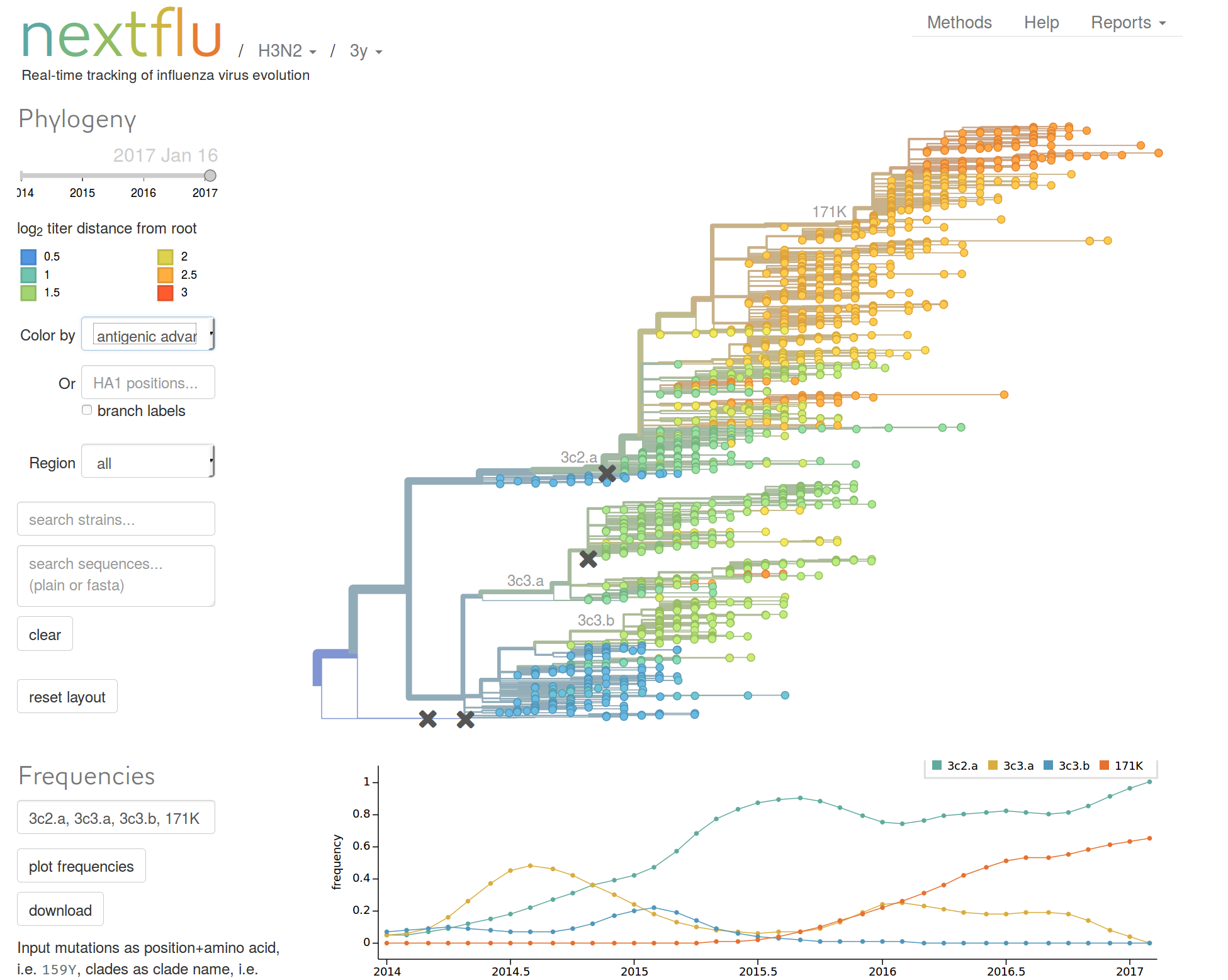
- Influenza virus evolves to avoid human immunity
- Vaccines need frequent updates

nextflu.org
joint work with Trevor Bedford & his lab
Real-time molecular epidemiology during outbreaks?
Ebola virus outbreak in West Africa -- Dudas et al, Nature 2017
How do we avoid the long delay?
nextstrain.org
joint work with Trevor Bedford & his lab
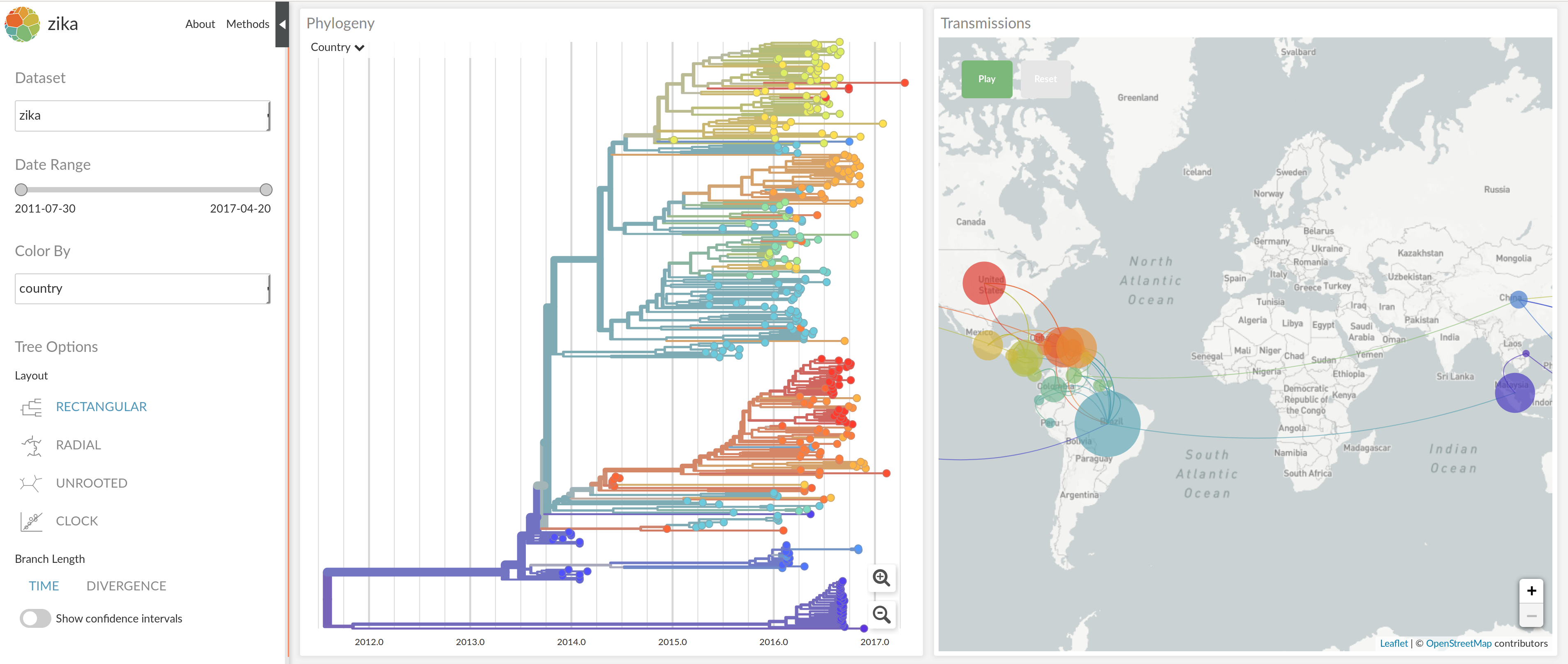
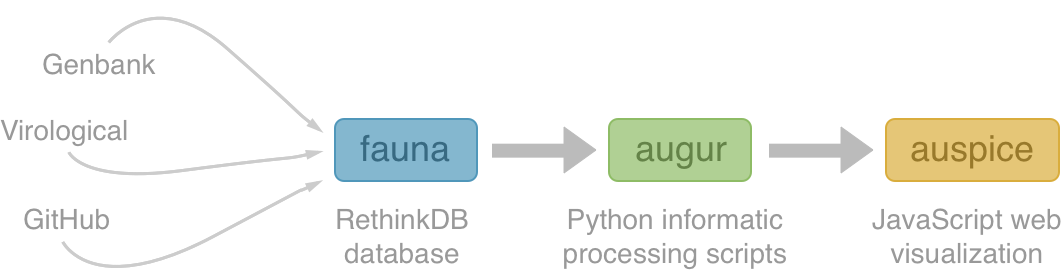
NextStrain architecture
Using treetime to rapidly compute timetrees
panX @ pangenome.de
S. pneumoniae data set by Croucher et al.
Barriers to data sharing: scientists
- Fear of being scooped/ensure maximal return
- Secondary analysis perceived as freeloading: "data parasites"
- Don't want to be second guessed
- Release and curation is laborious
- Sloppy records
Also see Smith et al, F1000 2016
Barriers to data sharing: organizations and governments
- Economic consequences of outbreaks (tourism, agriculture)
- Conflicts between high and low/middle income countries
- Concerns about IP and commercial exploitation
- Legislative/regulatory barriers
See also: van Panhius et al, BMC Public health 2014
Overcoming Barriers
Open vs restricted sharing
- Alternatives to GenBank: GenBank is "public domain", no requirement to credit data producers
- GISAID/EpiFlu: sign-up and agree to terms and conditions

- virological.org: Platform for sharing and discussing molecular epidemiology
- Explicit data reuse terms
- Outline planned projects in white-paper
- Caveat: Very difficult to enforce...
See Bogner et al, 2006
Building Trust
- Peter Bogner coordinated Influenza data sharing
- Andrew Rambaut coordinated Ebola virus data sharing
- During the EBV outbreak, WHO and journals explicitly encouraged data sharing
See also Smith et al, F1000 2016
Make sharing easy and provide incentives!
Challenges and Opportunities
- Software is not the bottleneck!
- Lack of data sharing
- timeliness of data sharing
- inconsistent formats
- Sharing is not a priority since nothing is done with data
- Sites like nextstrain provide incentives for data sharing
- Automated analysis and web dissemination are low maintenance
- Lots of synergies...
nextstrain.org







- Trevor Bedford
- Colin Megill
- Pavel Sagulenko
- Emma Hodcroft
- Sidney Bell
- James Hadfield
- Wei Ding




TreeTime & panX

TreeTime: Pavel Sagulenko
github.com/neherlab/treetimewebserver at treetime.ch
manuscript on bioRxiv

panX: Wei Ding
github.com/neherlab/pan-genome-analysislive site at pangenome.de
manuscript on bioRxiv