The evolutionary dynamics of untreated HIV and the maintenance of the latent reservoir
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/201802_dienstagsclub.html
Evolution of HIV
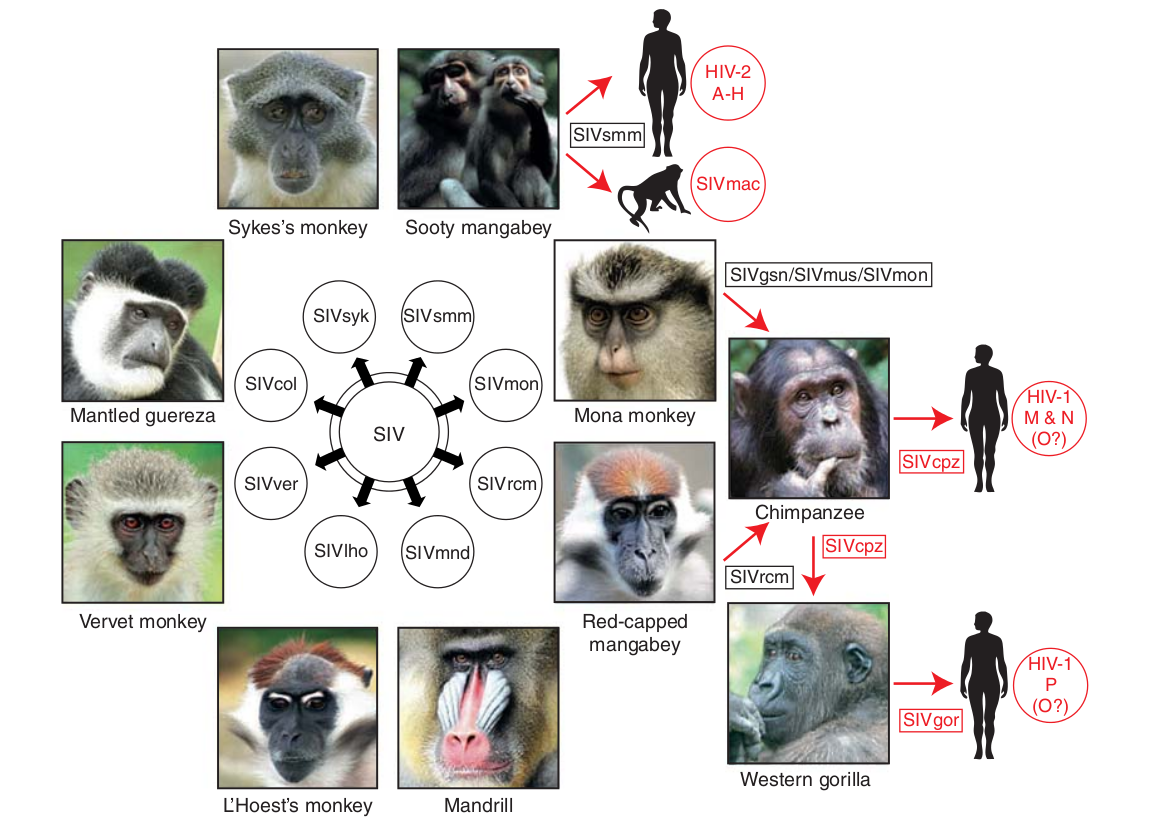
- Chimp → human transmission around 1900 gave rise to HIV-1 group M
- ~100 million infected people since
- subtypes differ at 10-20% of their genome
- HIV-1 evolves ~0.1% per year
HIV infection
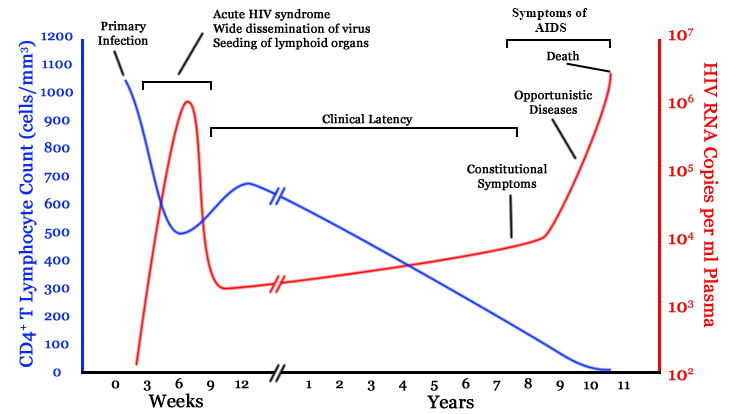
- $10^8$ cells are infected every day
- the virus repeatedly escapes immune recognition
- integrates into T-cells as
latent provirus
HIV-1 evolution within one individual

Accuracy of minor variant frequencies
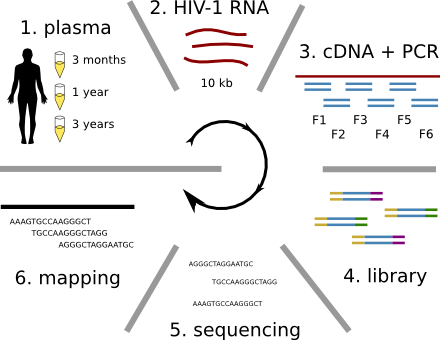
HIV-1 sequencing before and after therapy
 Zanini et al, eLife, 2015;
Brodin et al, eLife, 2016.
Collaboration with the group of Jan Albert
Zanini et al, eLife, 2015;
Brodin et al, eLife, 2016.
Collaboration with the group of Jan Albert
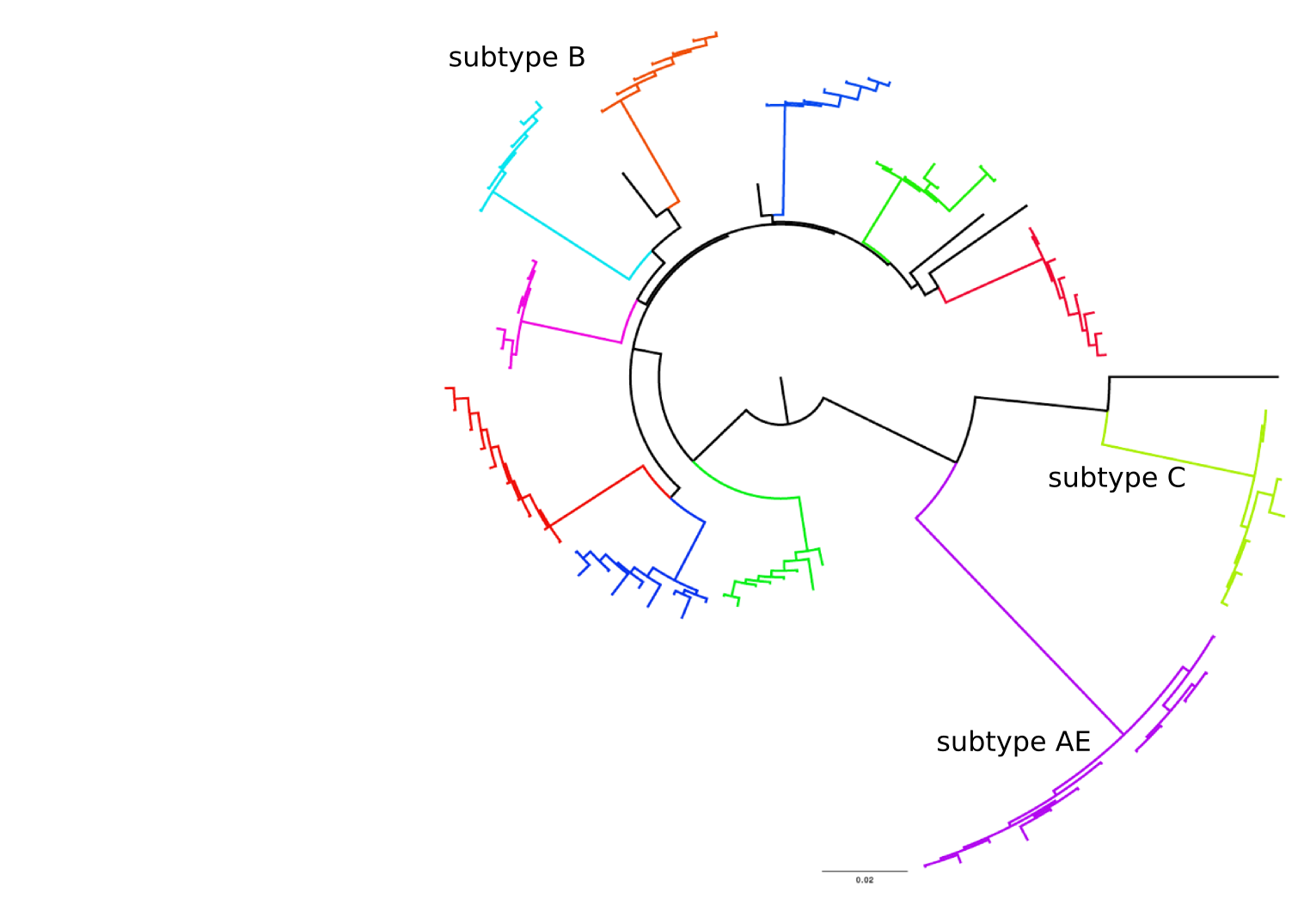
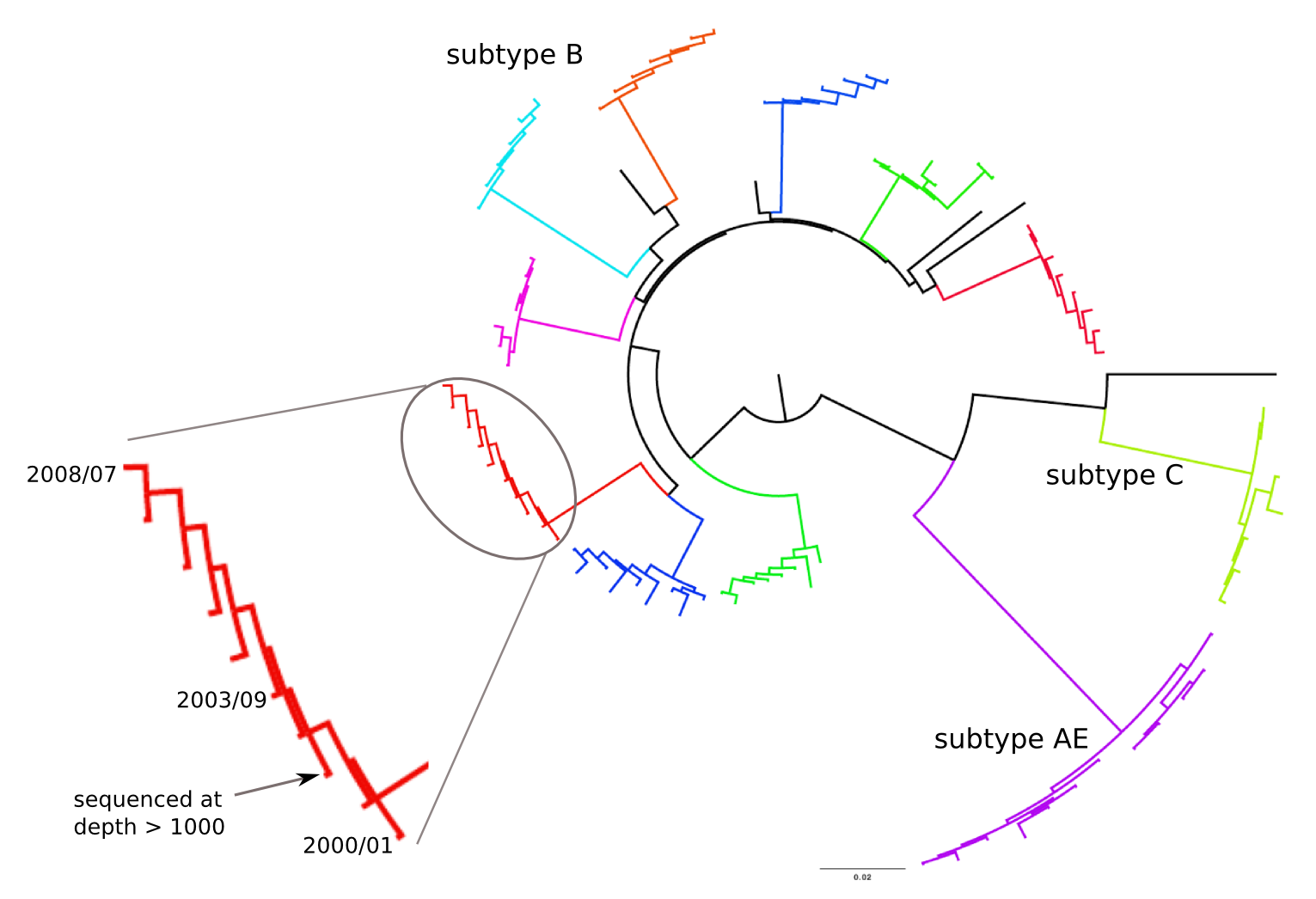
Population sequencing to track all mutations above 1%
- diverge at 0.1-1% per year
- almost full genomes coverage in 10 patients
- full data set at hiv.tuebingen.mpg.de
Mutation rates and diversity and neutral sites
Inferred vs measured mutations rates (Abram et al)
Diversity and rates of change
- envelope changes fastest, enzymes slowest
- identical rate of synonymous evolution
- diversity saturates where evolution is fast
- synonymous mutations stay at low frequency
Frequent reversion of previously beneficial mutations
- HIV escapes immune systems
- most mutations are costly
- humans selects for different mutations
- compensation or reversion?
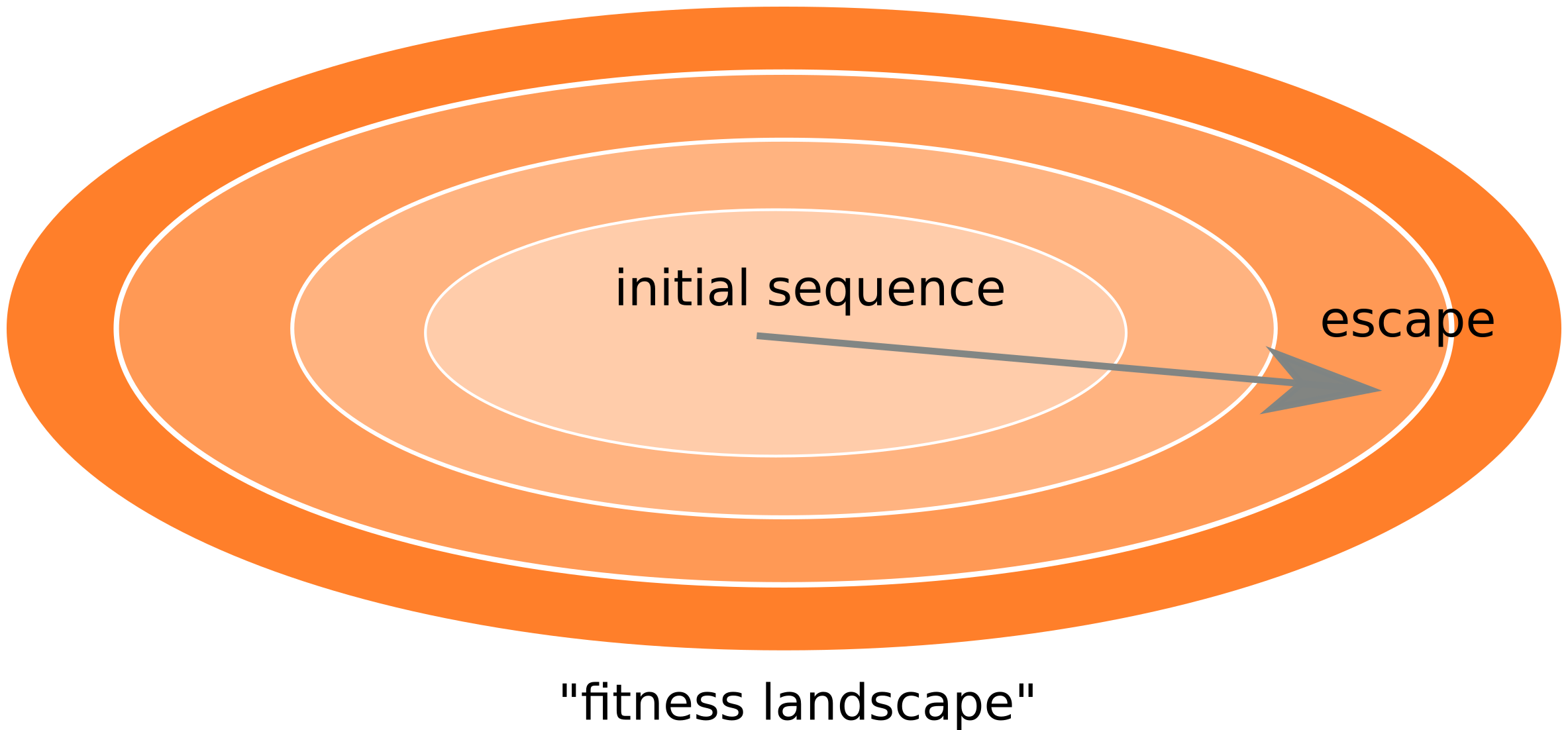
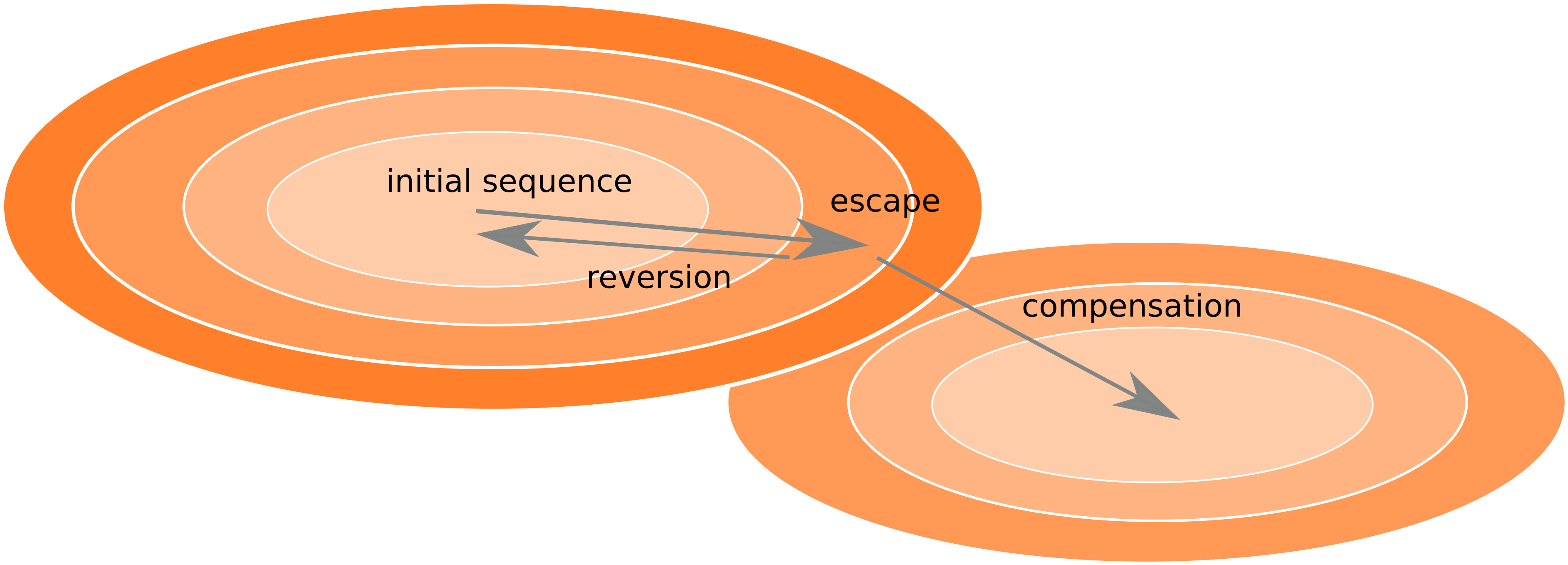
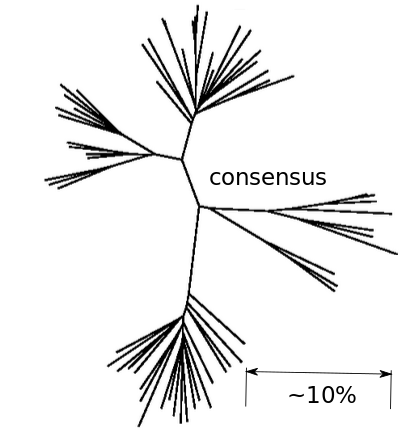
Inference of fitness costs
- mutation away from preferred state with rate $\mu$
- selection against non-preferred state with strength $s$
- variant frequency dynamics: $\frac{d x}{dt} = \mu -s x $
- equilibrium frequency: $\bar{x} = \mu/s $
- fitness cost: $s = \mu/\bar{x}$
Fitness landscape of HIV-1
Zanini et al, Virus Evolution, 2017Selection on RNA structures and regulatory sites
Zanini et al, Virus Evolution, 2017The distribution of fitness costs
Zanini et al, Virus Evolution, 2017Does HIV evolve during therapy?
 Brodin et al, eLife, 2016
Brodin et al, eLife, 2016
No evidence of ongoing replication
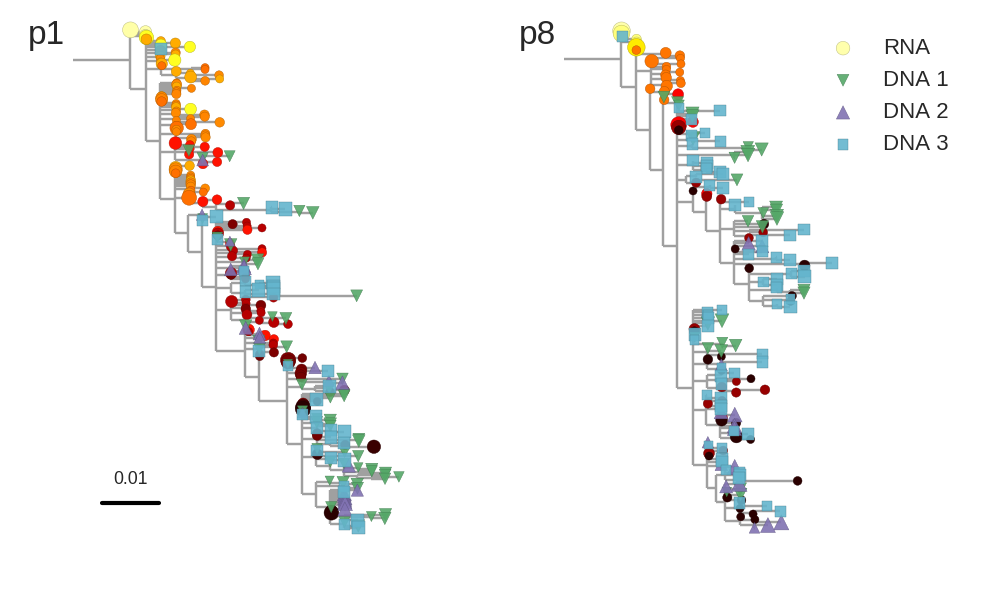
No evidence of ongoing replication
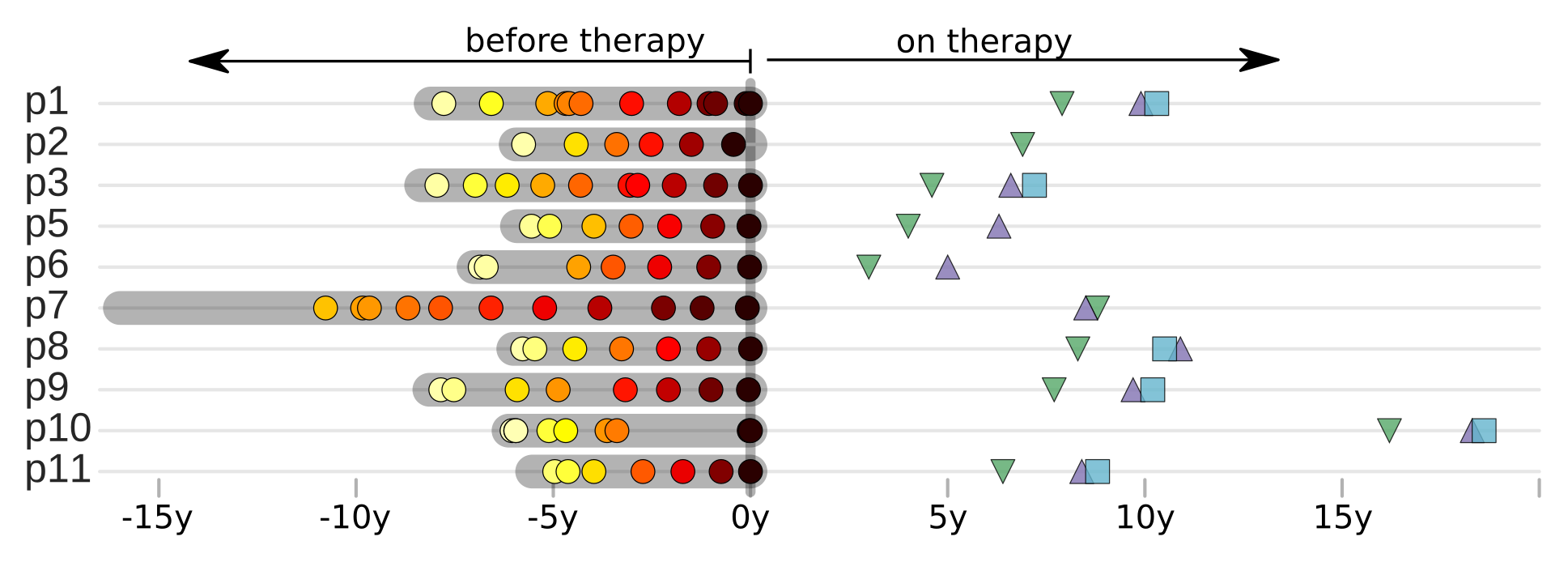
T-cell turnover is fast in untreated infection
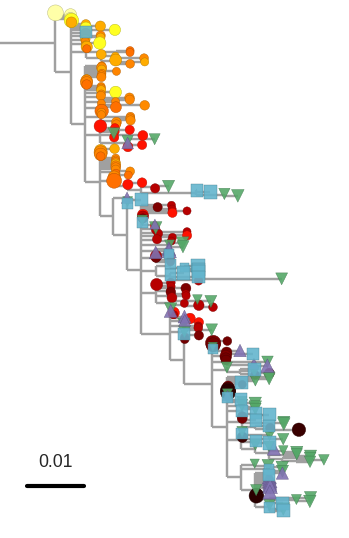
- latent HIV → barcode of a T-cell lineage
- all latent integrated virus derives from late infection
- untreated: T-cell lineages are short lived
- on therapy: T-cell clones live decades
Acknowledgments
- Fabio Zanini
- Jan Albert
- Johanna Brodin
- Christa Lanz
- Göran Bratt
- Lina Thebo
- Vadim Puller


