Technical meeting regarding nextstrain in Switzerland
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/201803_SSPAN.html
nextstrain.org
joint work with Trevor Bedford & his lab
code at github.com/nextstrain
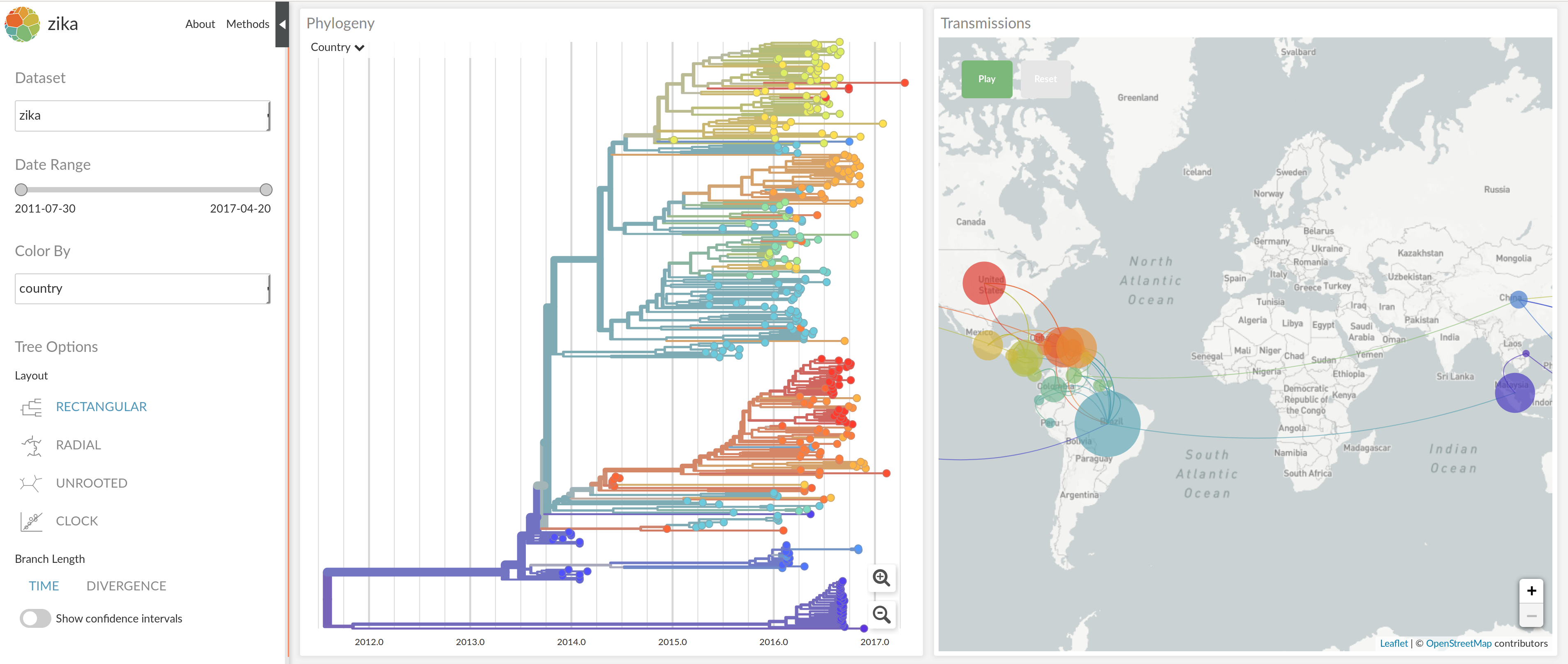
NextStrain architecture
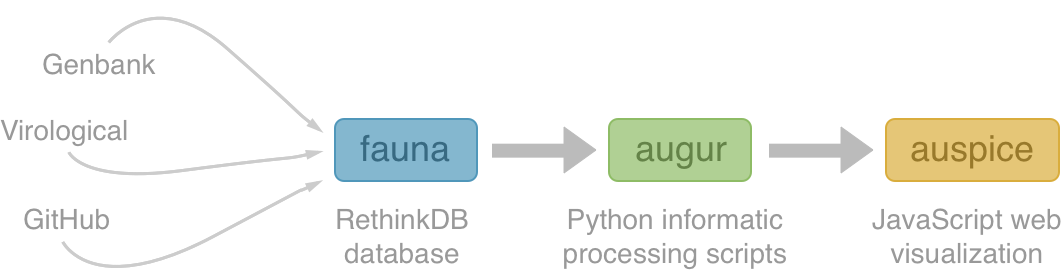
Using treetime to rapidly compute timetrees
TreeTime: maximum likelihood phylodynamic analysis
Phylogenetic trees record history:
- transmission
- divergence times
- population dynamics
- ancestral geographic distribution/migrations
Typical approach: Bayesian parameter estimation
- flexible
- probabilistic → confidence intervals etc
- but: computationally expensive
TreeTime by Pavel Sagulenko
- probabilistic treatment of divergence times
- dates trees with thousand sequences in a few minutes
- linear time complexity
- fixed tree topology
- github.com/neherlab/treetime
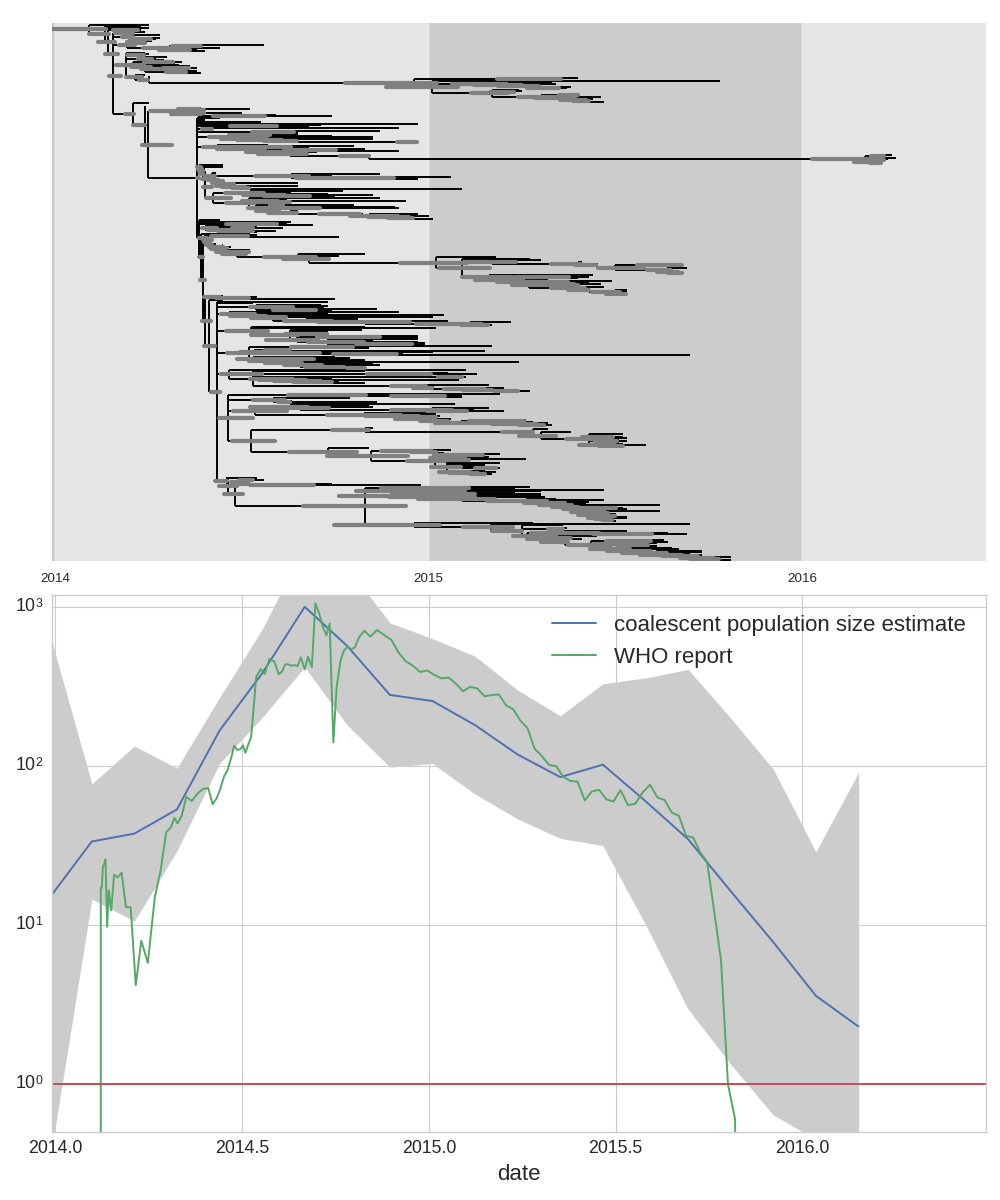
West African Ebola virus outbreak
TreeTime: nuts and bolts

Attach sequences and dates

Reconstruct ancestral sequences
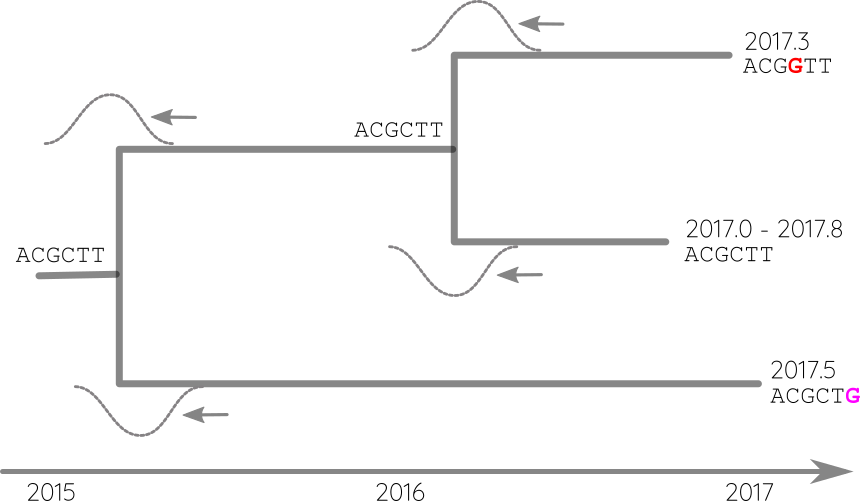
Propagate temporal constraints via convolutions
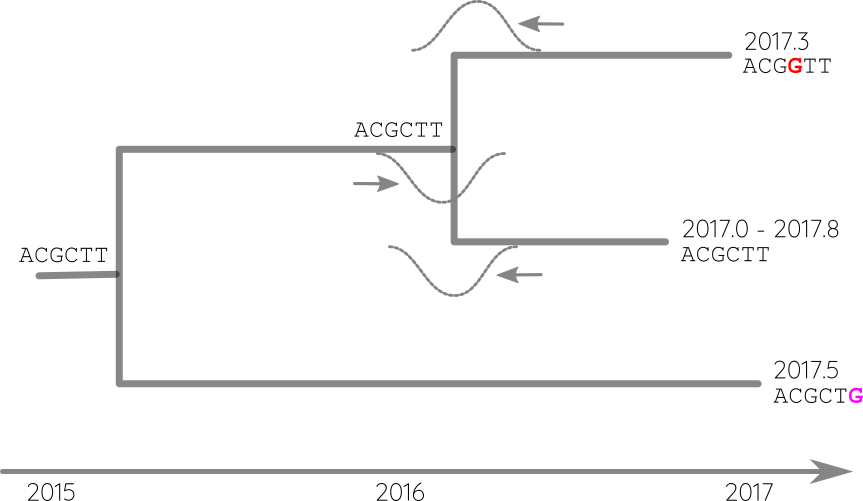
Integrate up-stream and down-stream constraints
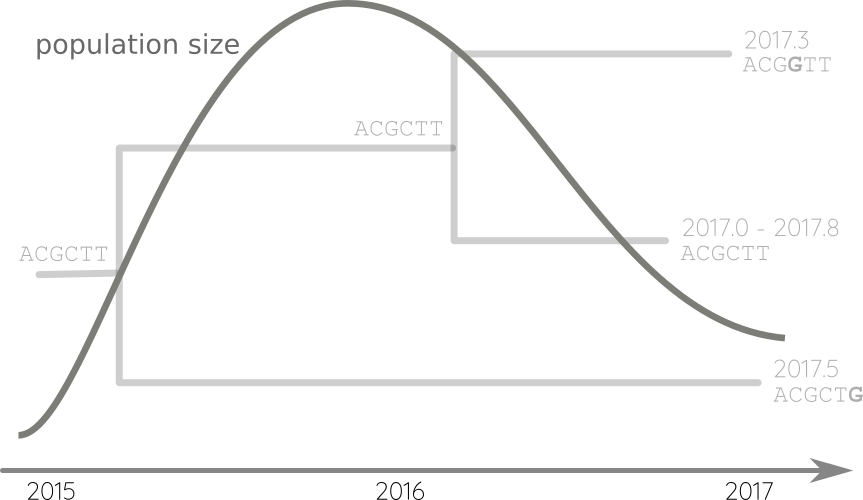
Fit phylodynamic model → iterate
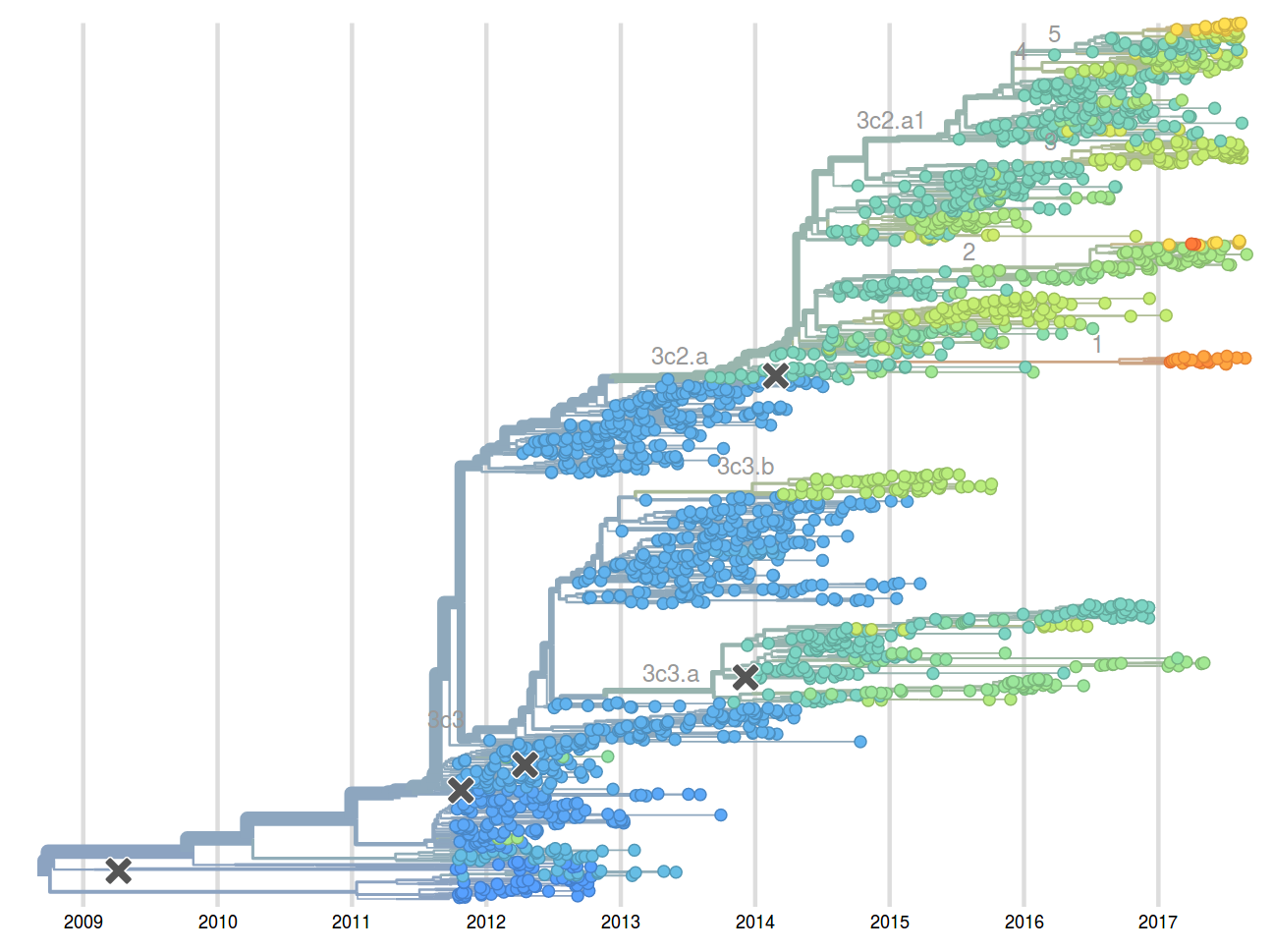
Molecular clock phylogenies of ~2000 A/H3N2 HA sequences -- a few minutes
What about bacteria?
- vertical and horizontal transmission
- genome rearrangements
- much larger genomes
- variation of divergence along the genome
- NGS genomes tend to be fragmented
- annotations of variable quality

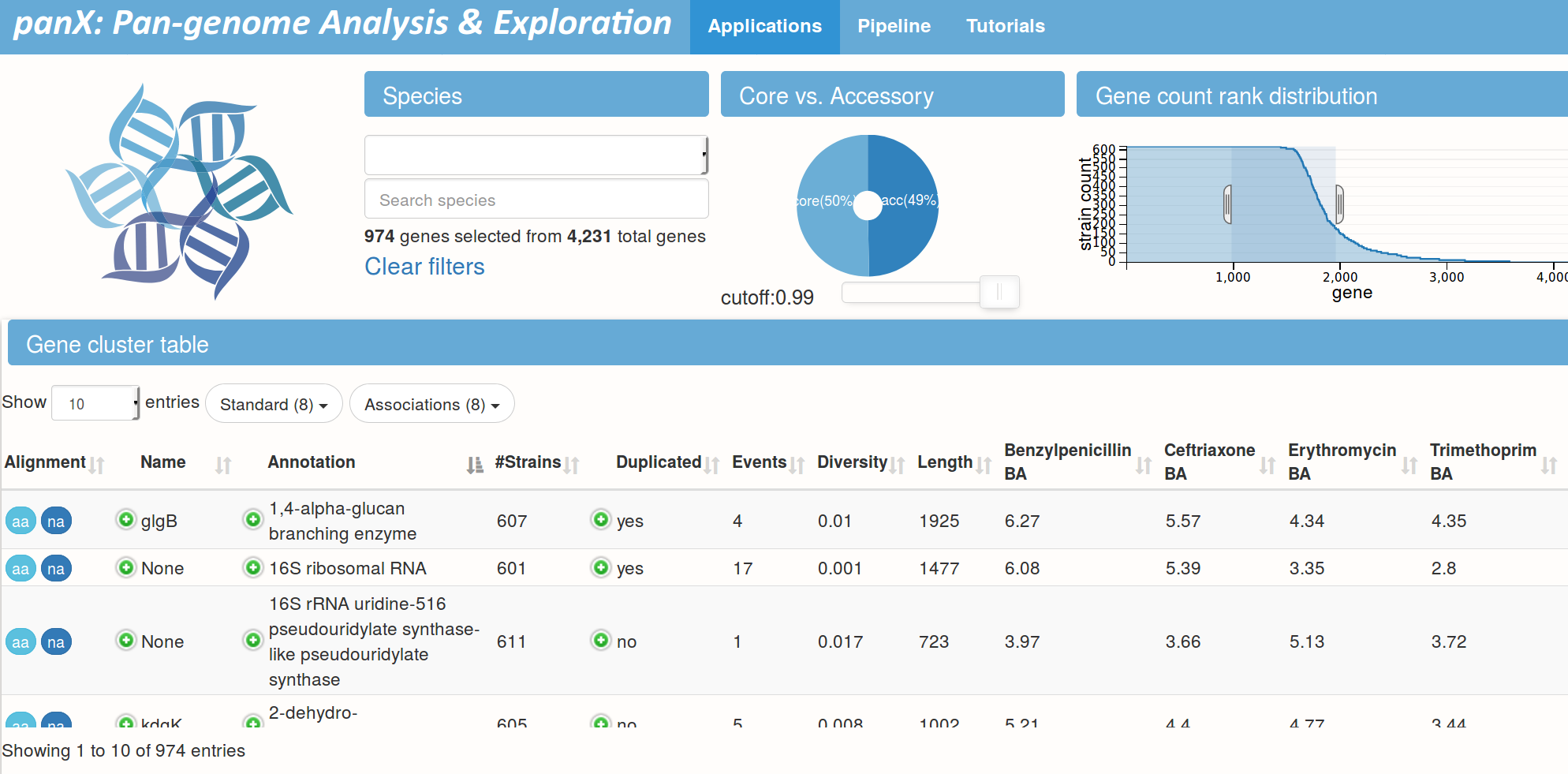
panX by Wei Ding
- pan-genome identification pipeline
- phylogenetic analysis of each orthologous cluster
- detect associations with phenotypes
- fast: analyze hundreds of genomes in a few hours
- github.com/neherlab/pan-genome-analysis
panX @ pangenome.de
S. pneumoniae data set by Croucher et al.
Pan-genome statistics and filters
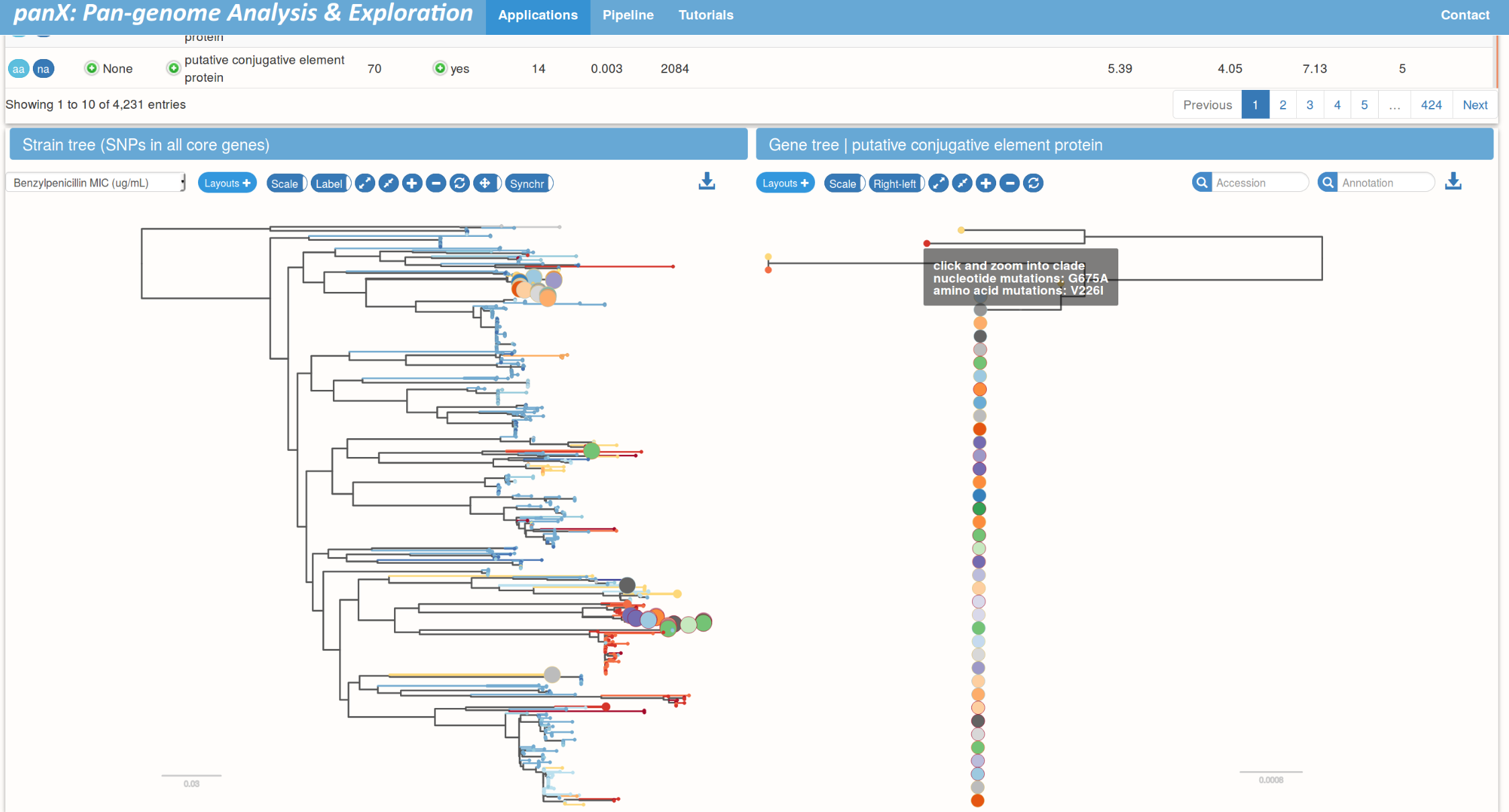
Species trees and gene trees
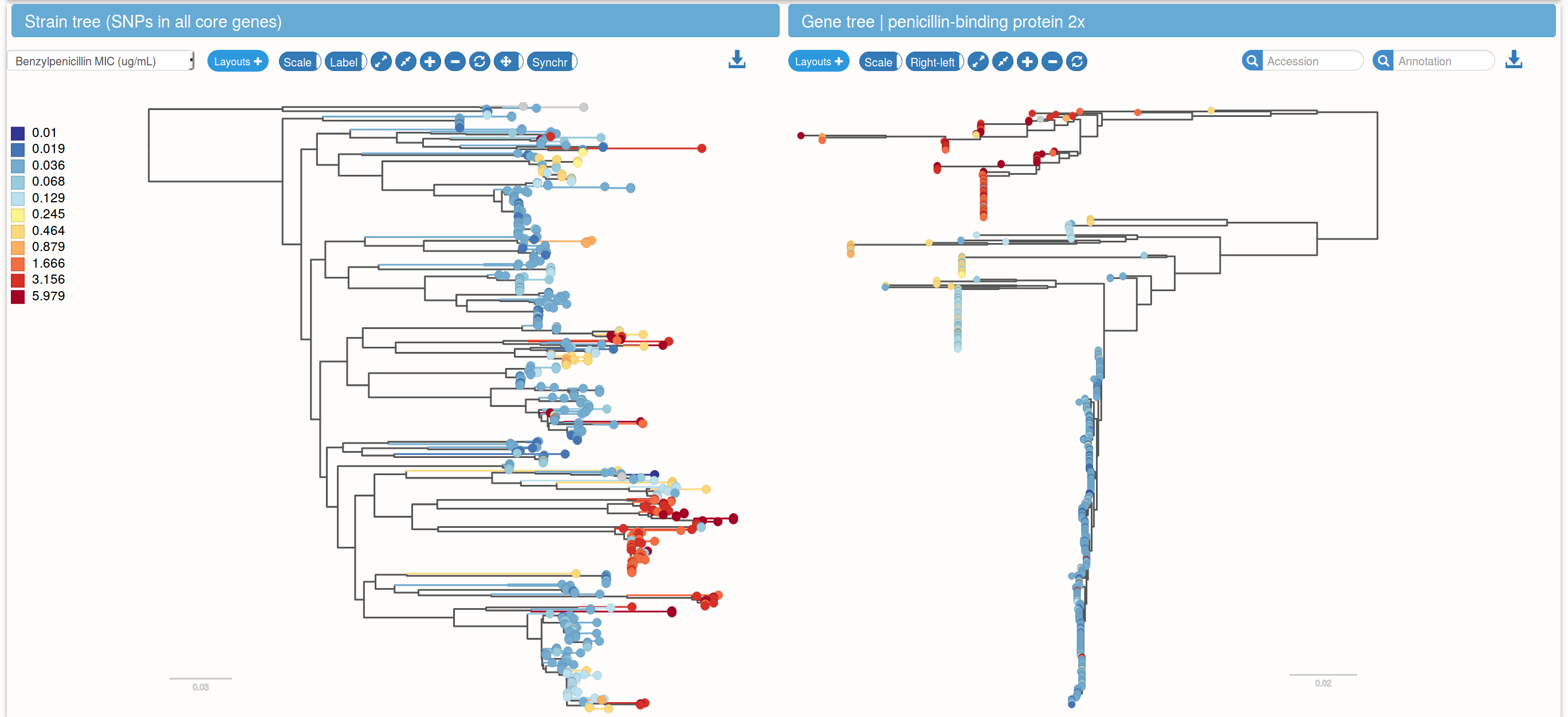
Links between species trees and gene trees
Processing for nextstrain
- Python 2.7, raxml/iqtree/fasttree, MAFFT, TreeTime
- run time of approx one hour, 1-4CPUs, limited by tree building
- Sequences: for viruses we use fasta, for bacteria vcf
- Metadata: best as tsv or json
- Ideally we would want some hybrid between nextstrain and panX for bacteria
Processing for panX
- Python 2.7, raxml/fasttree, DIAMOND, MCL, MAFFT, TreeTime
- run time of approx six hours, 64CPUs, limited by all-against-all comparison and tree building for every gene
- Sequences: genbank files with annotation, gff could be used as well
- Metadata: best as tsv
Visualization: nextstrain
- runs a node server, but basically a static site
- based on react and a custom tree viz (phylotree)
- we will need authentification
- currently uses mapbox/leaflet maps (would need an API key)
nextstrain.org




- Trevor Bedford
- Colin Megill
- Sidney Bell
- James Hadfield
- All the scientist that share virus sequence data




TreeTime & panX

TreeTime: Pavel Sagulenko
github.com/neherlab/treetimewebserver at treetime.ch
manuscript on bioRxiv

panX: Wei Ding
github.com/neherlab/pan-genome-analysislive site at pangenome.de
manuscript on bioRxiv



