Ist die Evolution von Grippeviren vorhersehbar?
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/201803_SeniorenUni.html
Viruses
 tobacco mosaic virus
(Thomas Splettstoesser, wikipedia)
tobacco mosaic virus
(Thomas Splettstoesser, wikipedia)
bacteria phage (adenosine, wikipedia)
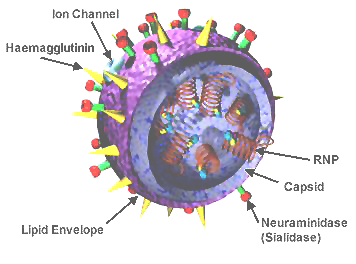
influenza virus wikipedia
human immunodeficiency virus (HIV) wikipedia
- Viren benötigen Wirtszellen zur Replikation
- sind wenig mehr als Genom + Kapsid
- ihr Genom ist 5-200Tsd Basen lang (+Ausnahmen)
- zahlenmäßig dominierenden Organismen $\sim 10^{31}$
Influenza virus
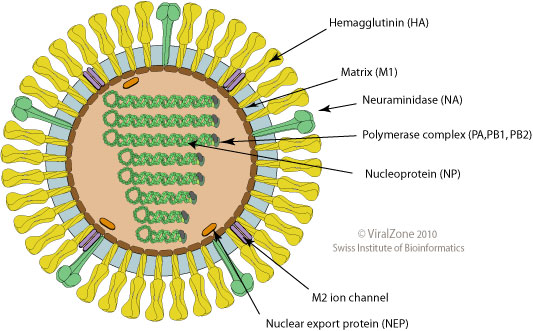
- Surface proteins hemagglutinin (HA) and neuraminidase (NA)
- Influenza A virus
- Common in birds and mammals
- Many different subtypes defined by surface proteins
- H3N2, H1N1, H7N9, H5N1
- Influenza B virus
- infects mainly humans
- two lineages that split 30-40y ago
- B/Victoria vs B/Yamagata
Human Influenza A viruses

Weekly numbers of positive influenza tests in the US by subtype
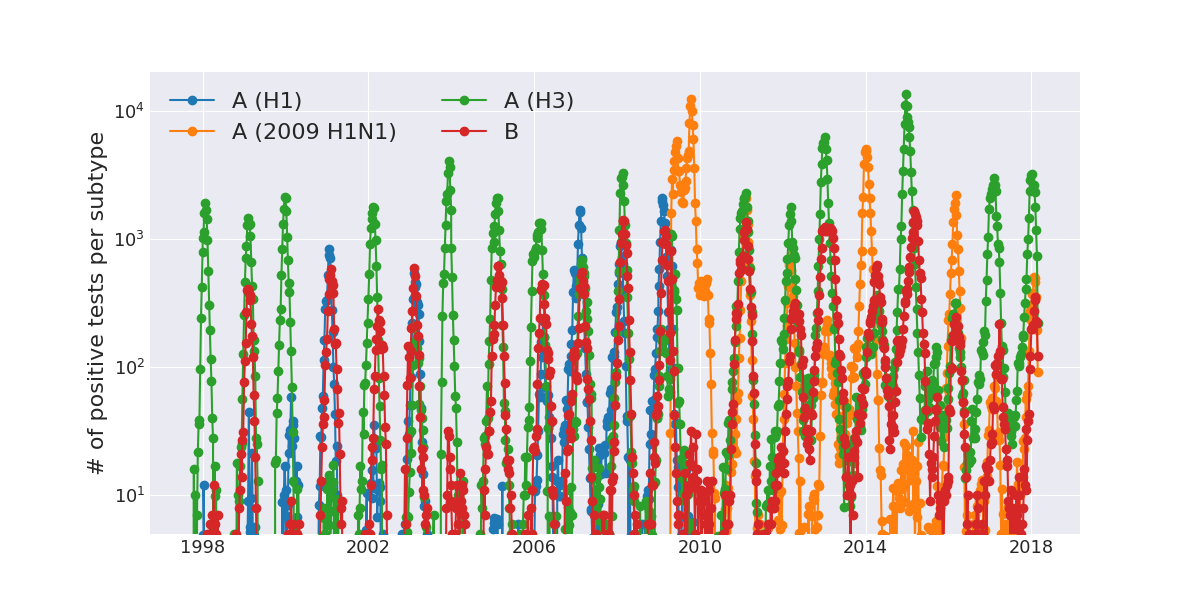 Data by the US CDC
Data by the US CDC
Tracking virus spread and evolution by sequencing
A/Brisbane/100/2014
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
A/Brisbane/1000/2015
GGATAATTCTATTAACCATGAAGACTATTATTGCTTT...
A/Brisbane/1/2017
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
... thousands of sequences...
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
A/Brisbane/1000/2015
GGATAATTCTATTAACCATGAAGACTATTATTGCTTT...
A/Brisbane/1/2017
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
... thousands of sequences...
Phylogenetic analysis of viral sequences
RNA viruses have a high mutation rate. New mutations arise every few weeks.



- Influenza virus evolves to avoid human immunity
- Vaccines need frequent updates
nextflu.org
joint work with Trevor Bedford & his lab
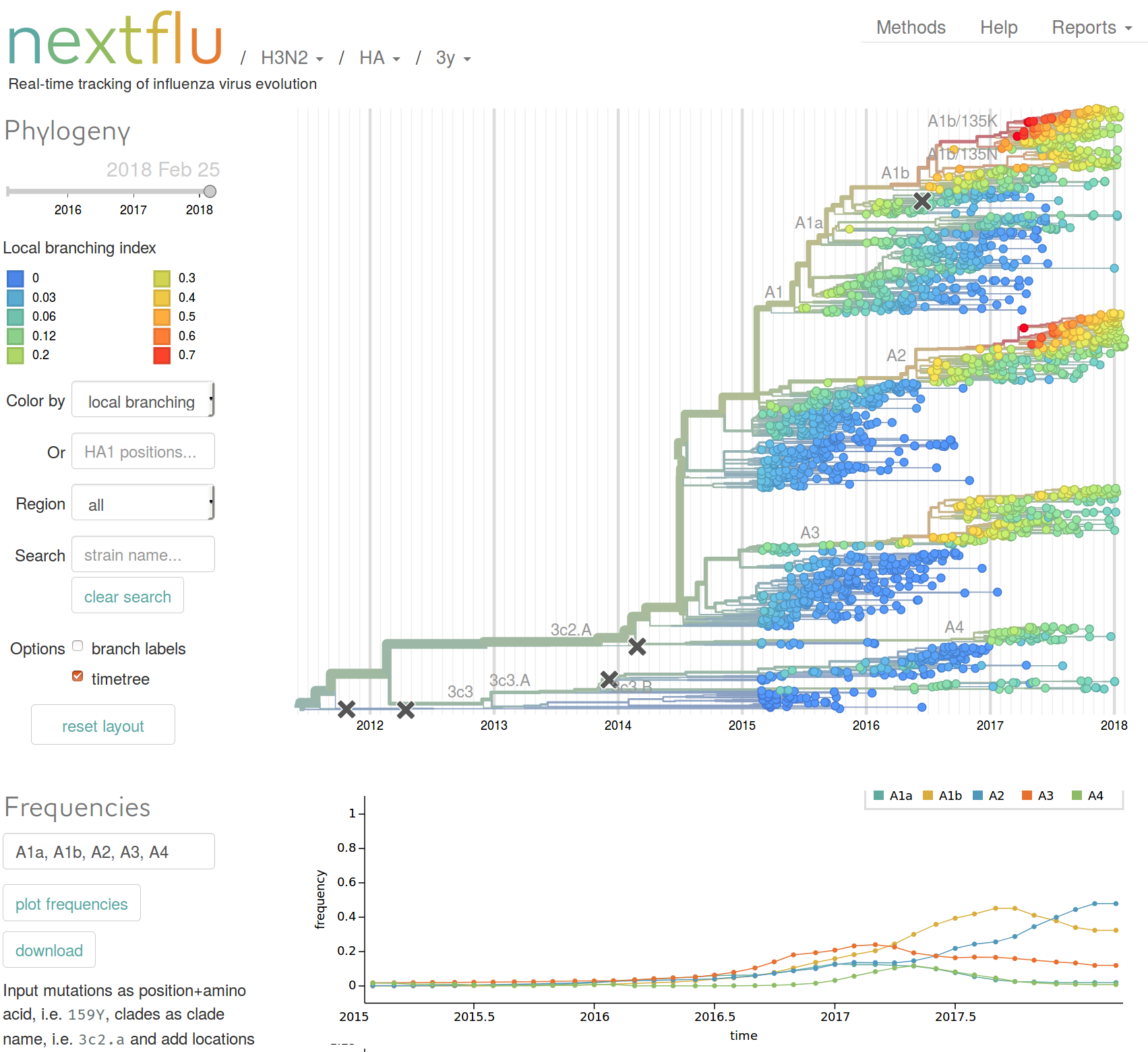
Can we predicting influenza virus evolution?

- Extrapolate current trends
- Predict based on historical patterns
- Use the branching pattern to predict success
- Measure antigenic properties of circulating viruses
Fitness inference from trees
$$P(\mathbf{x}|T) = \frac{1}{Z(T)} p_0(x_0) \prod_{i=0}^{n_{int}} g(x_{i_1}, t_{i_1}| x_i, t_i)g(x_{i_2}, t_{i_2}| x_i, t_i)$$
RN, Russell, Shraiman, eLife, 2014
Prediction of the dominating H3N2 influenza strain
- no influenza specific input
- how can the model be improved? (see model by Luksza & Laessig)
- what other context might this apply?
nextstrain.org
joint work with Trevor Bedford & his lab
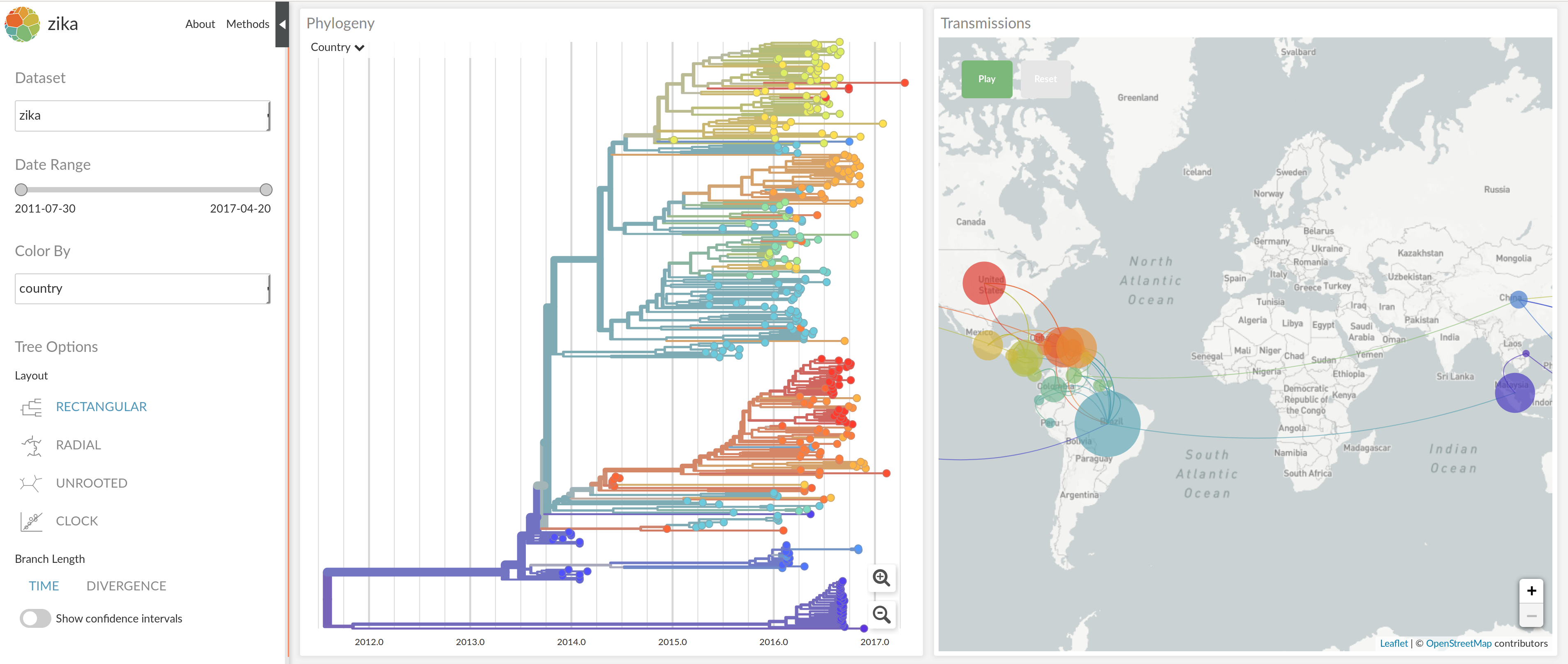
Summary
- RNA viruses evolve fast
- Viral lineages pick up mutations every few weeks
- Genomic sequences can be used to track viral spread
- Algorithms can predict future influenza virus variants
- Automated real-time analysis can help fight the spread of disease
Joint work with....



- Boris Shraiman
- Colin Russell
- Trevor Bedford



nextstrain.org






- Trevor Bedford
- Colin Megill
- Pavel Sagulenko
- Sidney Bell
- James Hadfield
- Wei Ding



