Real-time phylodynamics with nextflu and nextstrain
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/201806_BSSE.html
How this all started...



.... population genetics of rapid adaptation and predicting flu




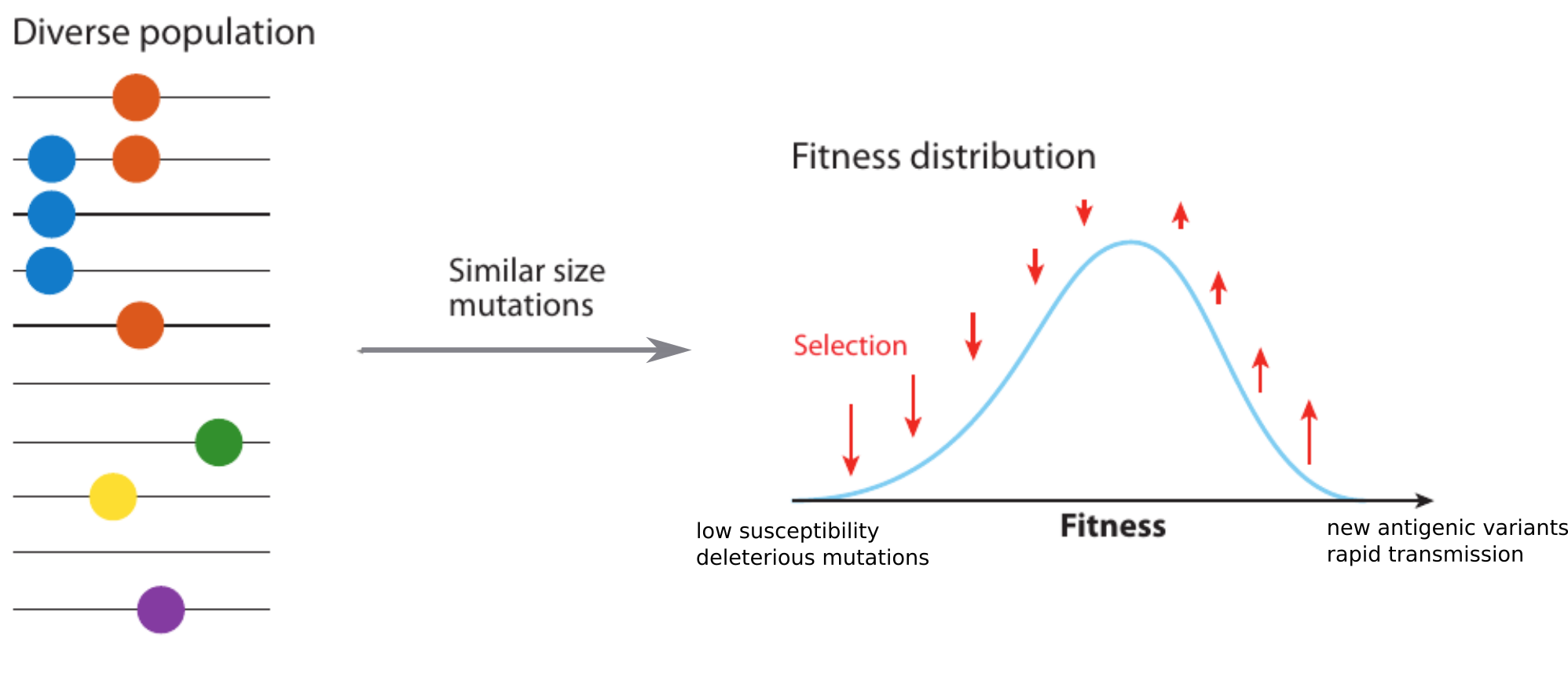
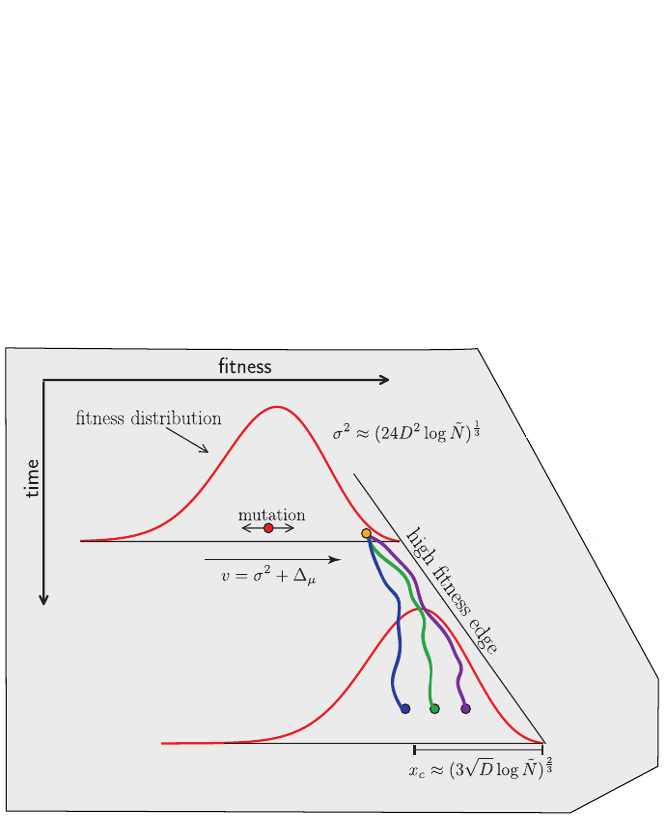
Model of rapidly adapting virus populations
Typical tree

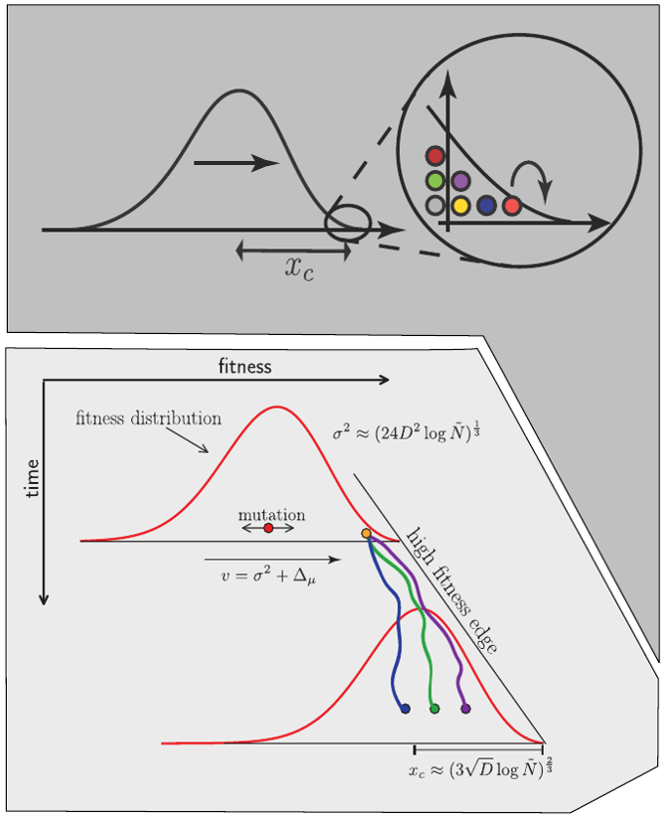
Bolthausen-Sznitman Coalescent
Bursts in a tree ↔ high fitness genotypes
Predicting evolution

Given the branching pattern:
- can we predict fitness?
- pick the closest relative of the future?
Fitness inference from trees
$$P(\mathbf{x}|T) = \frac{1}{Z(T)} p_0(x_0) \prod_{i=0}^{n_{int}} g(x_{i_1}, t_{i_1}| x_i, t_i)g(x_{i_2}, t_{i_2}| x_i, t_i)$$
$$\partial_t g( x,t'|y, t) = [y - 2\phi_{\omega} (y,t)] g(x,t'| y,t)-\sigma^2 \partial_yg( x,t'| y, t) +D \partial_y^2 g( x,t'|y,t)$$
RN, Russell, Shraiman, eLife, 2014
Validation on simulated data
nextstrain.org/flu
joint work with Trevor Bedford & his lab
code at github.com/nextstrain
NextStrain architecture
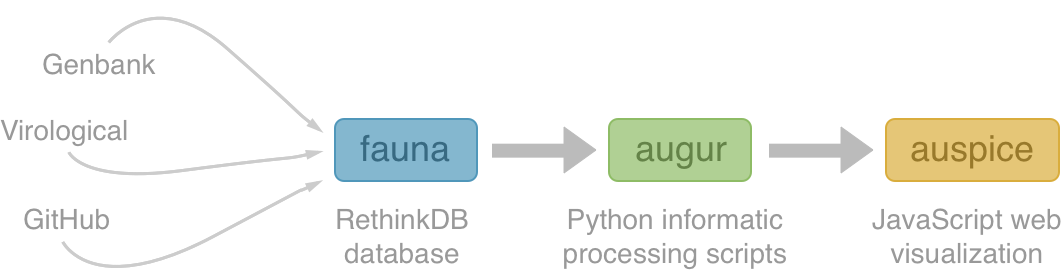
Using treetime to rapidly compute timetrees
TreeTime: maximum likelihood phylodynamic analysis
desired features:
- ancestral sequences
- divergence times
- ancestral geographic distribution
- population dynamics
Typical approach: Bayesian parameter estimation
- flexible
- probabilistic → confidence intervals etc
- but: computationally expensive
TreeTime by Pavel Sagulenko
- probabilistic treatment of divergence times
- dates trees with thousand sequences in a few minutes
- linear time complexity
- fixed tree topology
- github.com/neherlab/treetime
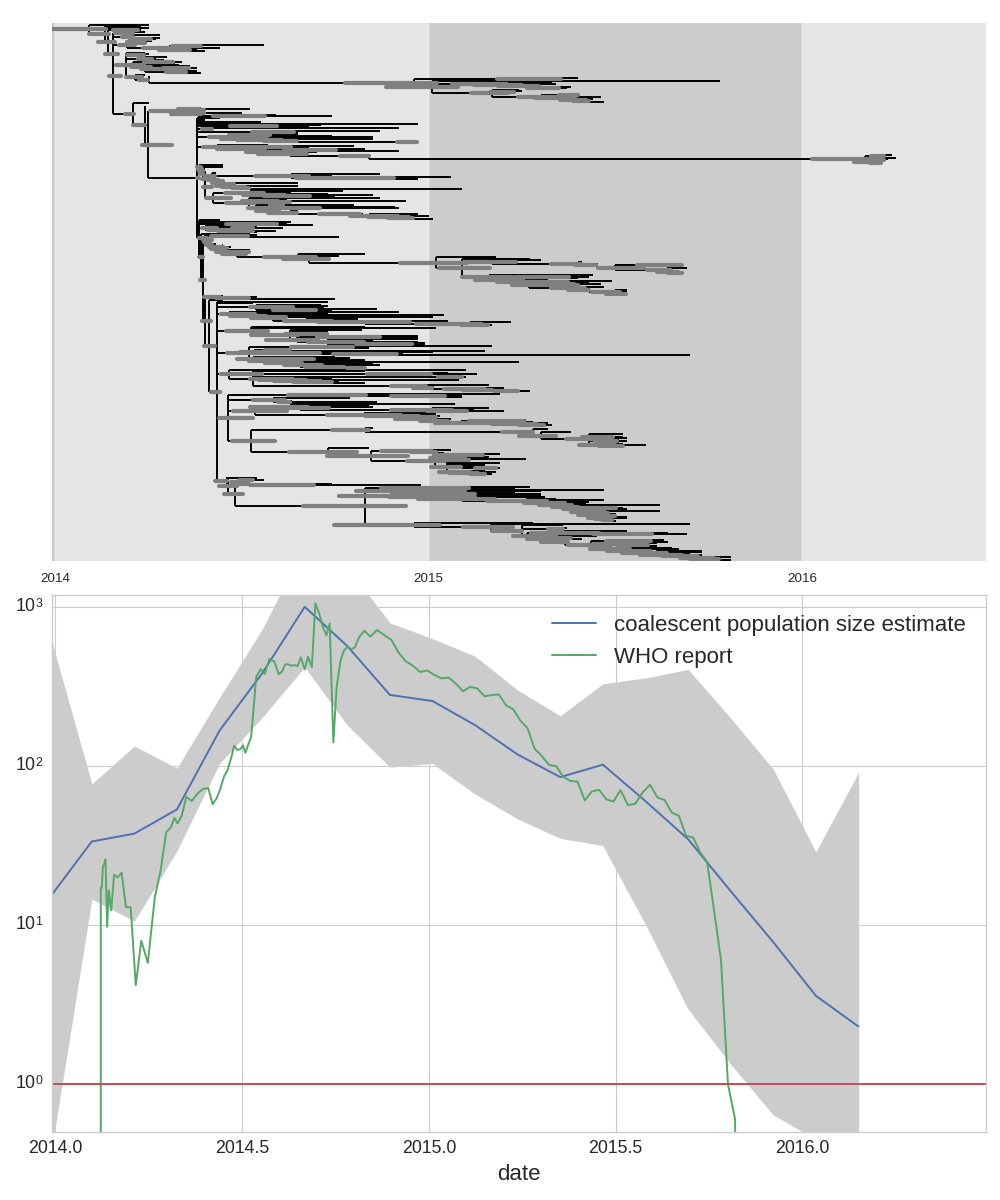
West African Ebola virus outbreak
TreeTime: nuts and bolts

Attach sequences and dates

Reconstruct ancestral sequences
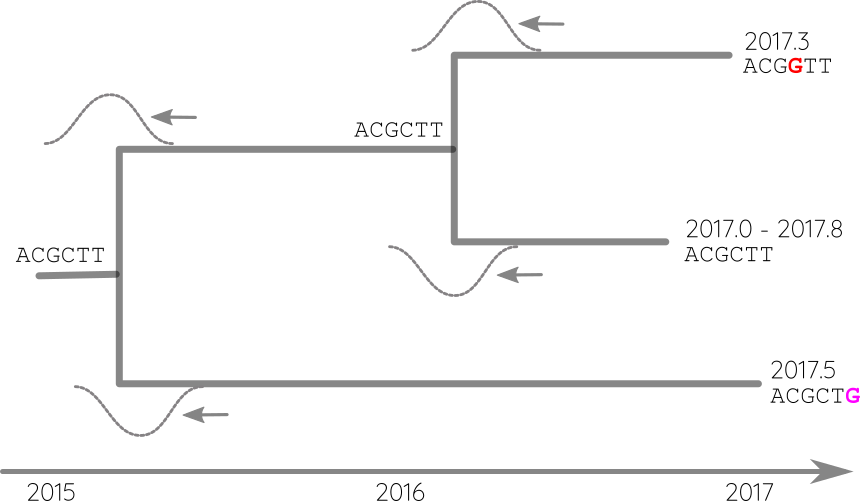
Propagate temporal constraints via convolutions
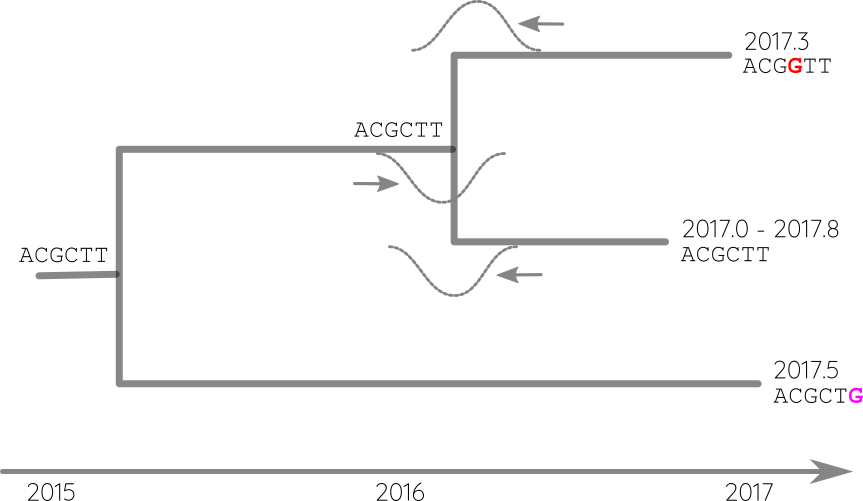
Integrate up-stream and down-stream constraints
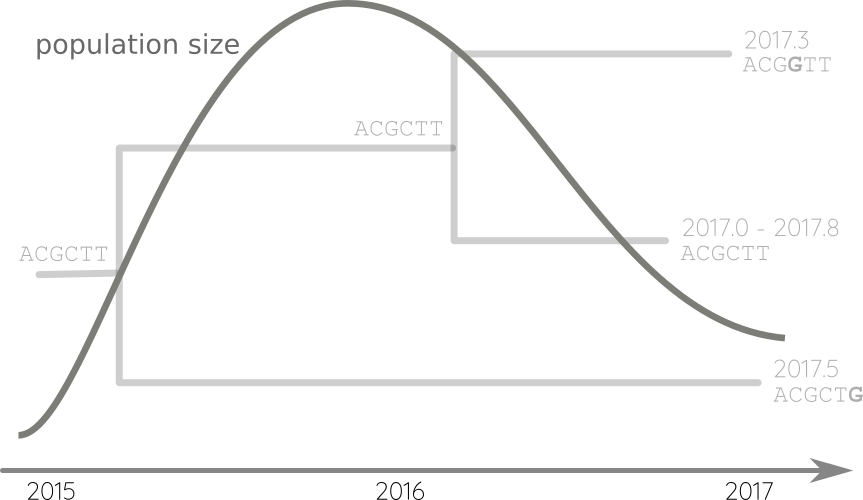
Fit phylodynamic model → iterate
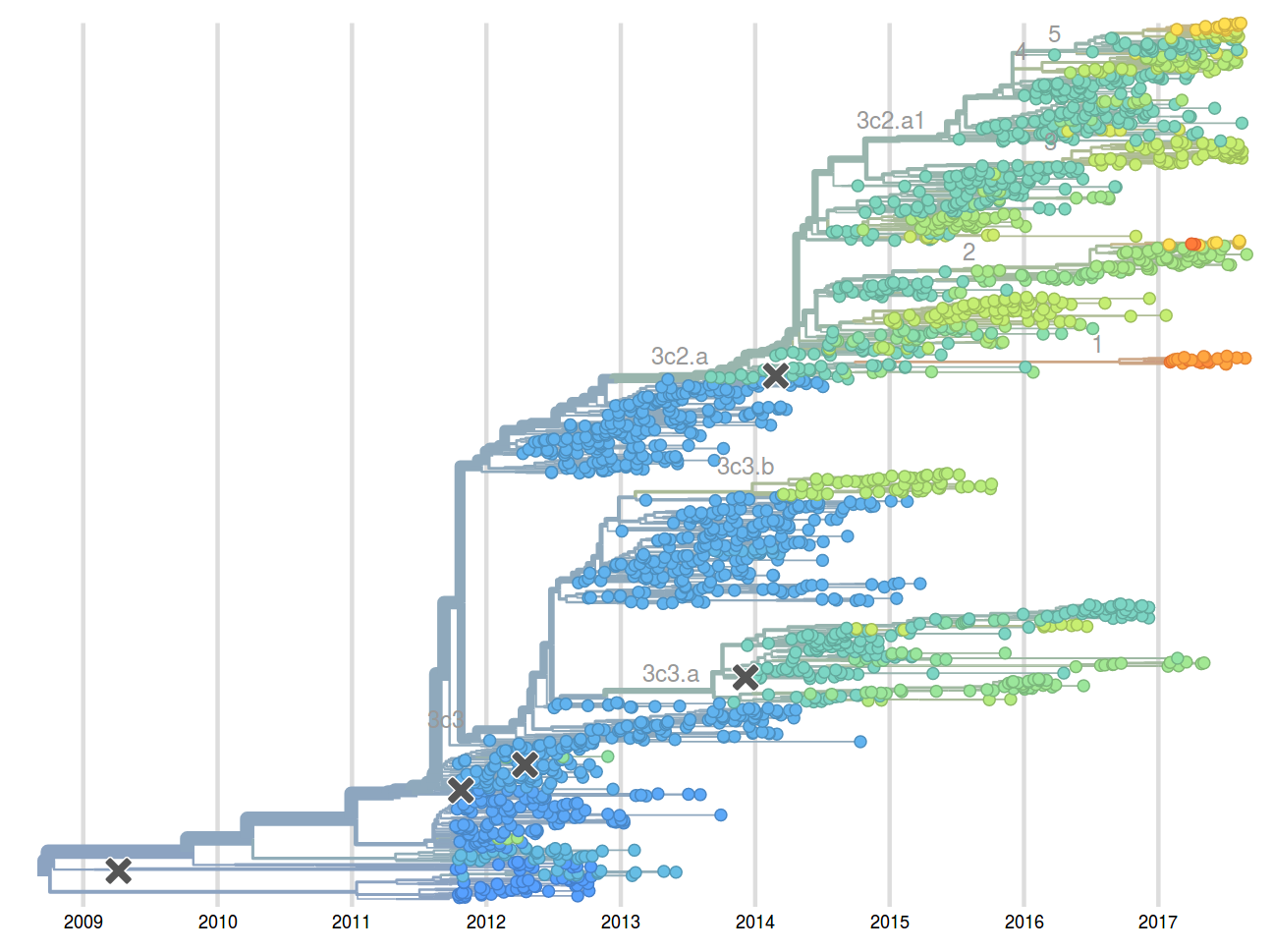
Molecular clock phylogenies of ~2000 A/H3N2 HA sequences -- a few minutes
Integrating phenotype and genotype data
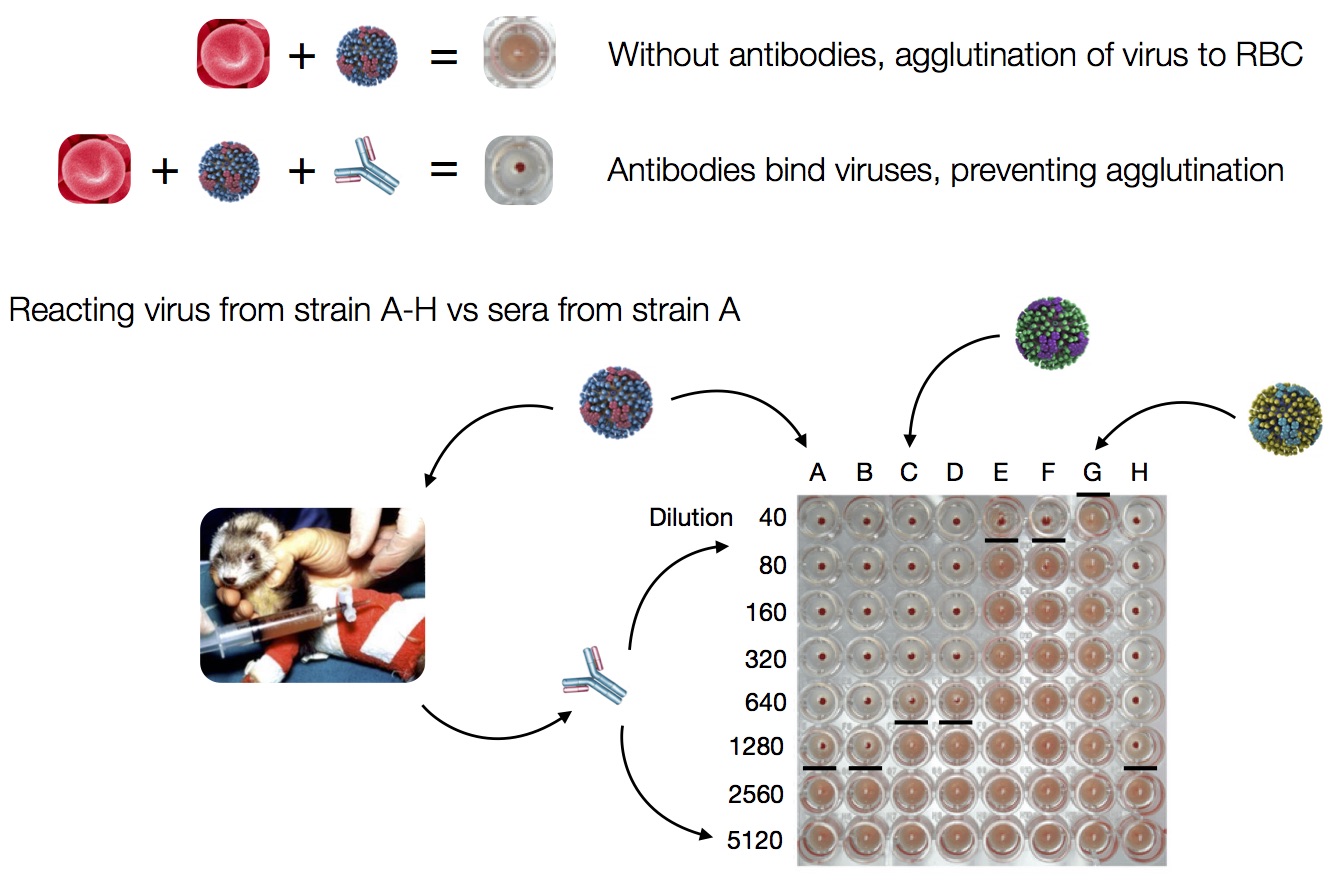
Antigenic distance tables
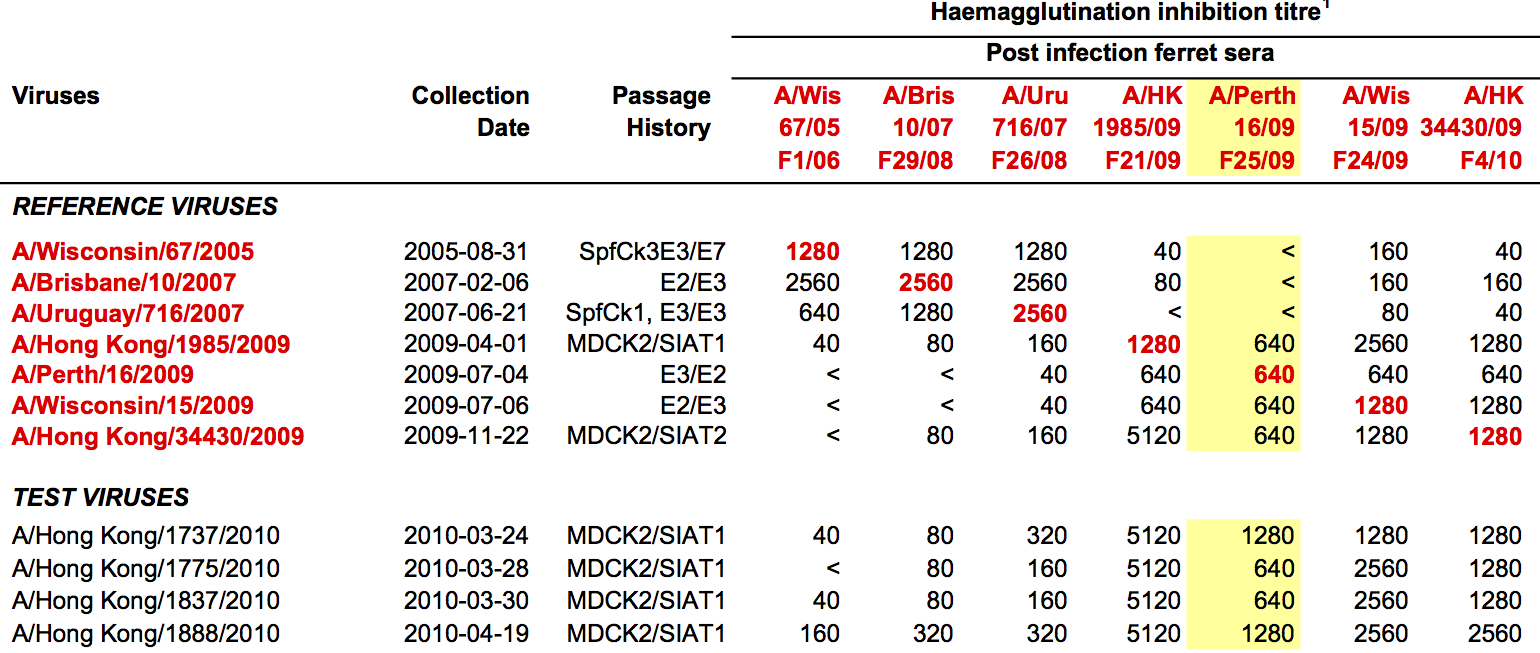
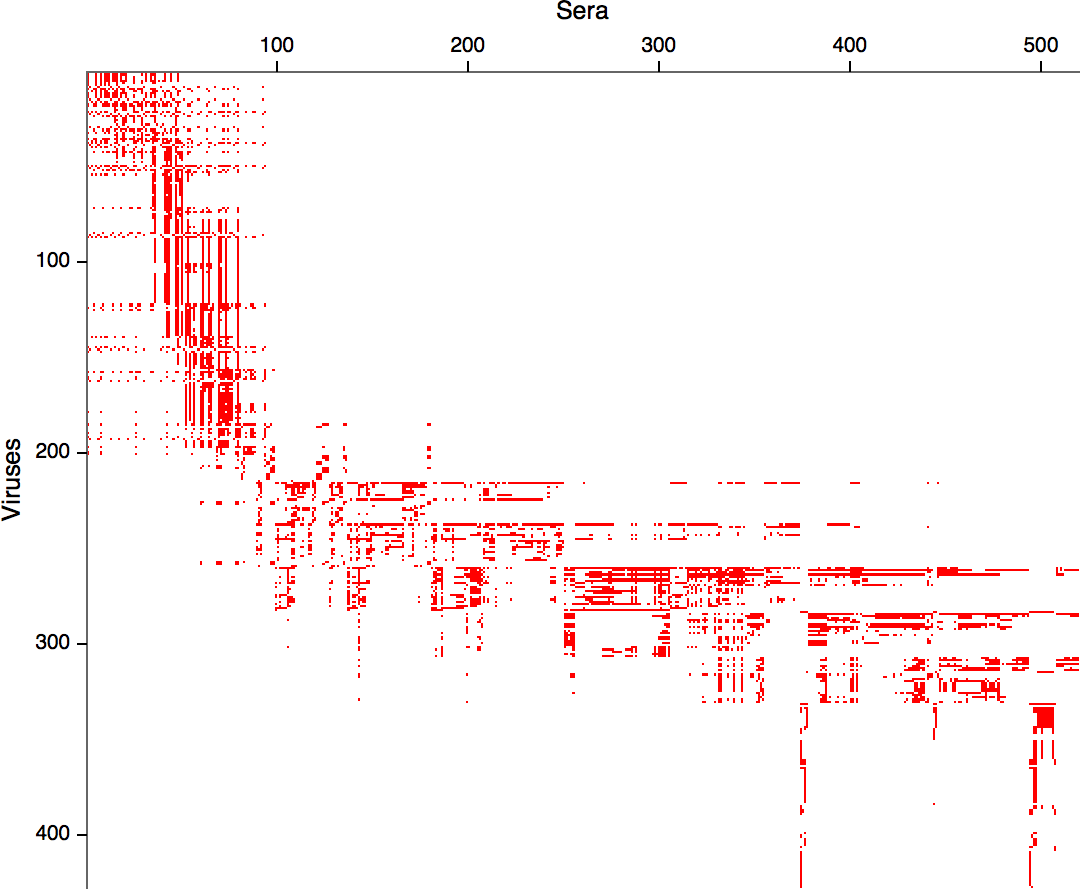
- Long list of distances between sera and viruses
- Tables are sparse, only close by pairs
- Structure of space is not immediately clear
- MDS in 2 or 3 dimensions
Integrating antigenic and molecular evolution
- each branch contributes $d_i$ to antigenic distance
- sparse solution for $d_i$ through $l_1$ regularization
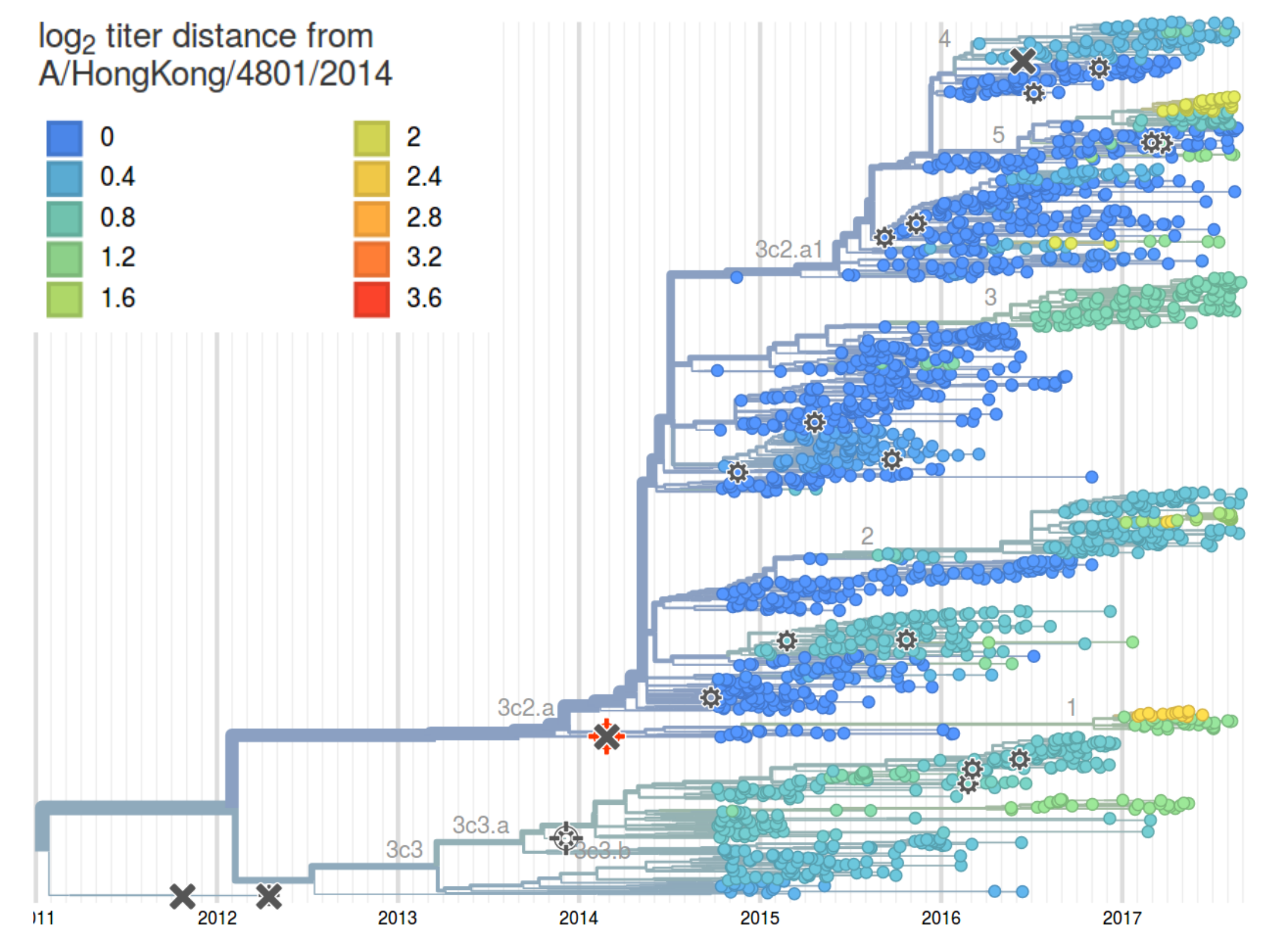
HI distances on the phylogenetic tree
Acknowlegdements






- Trevor Bedford
- Colin Megill
- Pavel Sagulenko
- Sidney Bell
- James Hadfield
- Wei Ding
- Emma Hodcroft
- Sanda Dejanic



