Using open data to track and predict infectious disease
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/201809_OpenCon.html
Data sharing in public health emergencies
data owner ↔ public interest
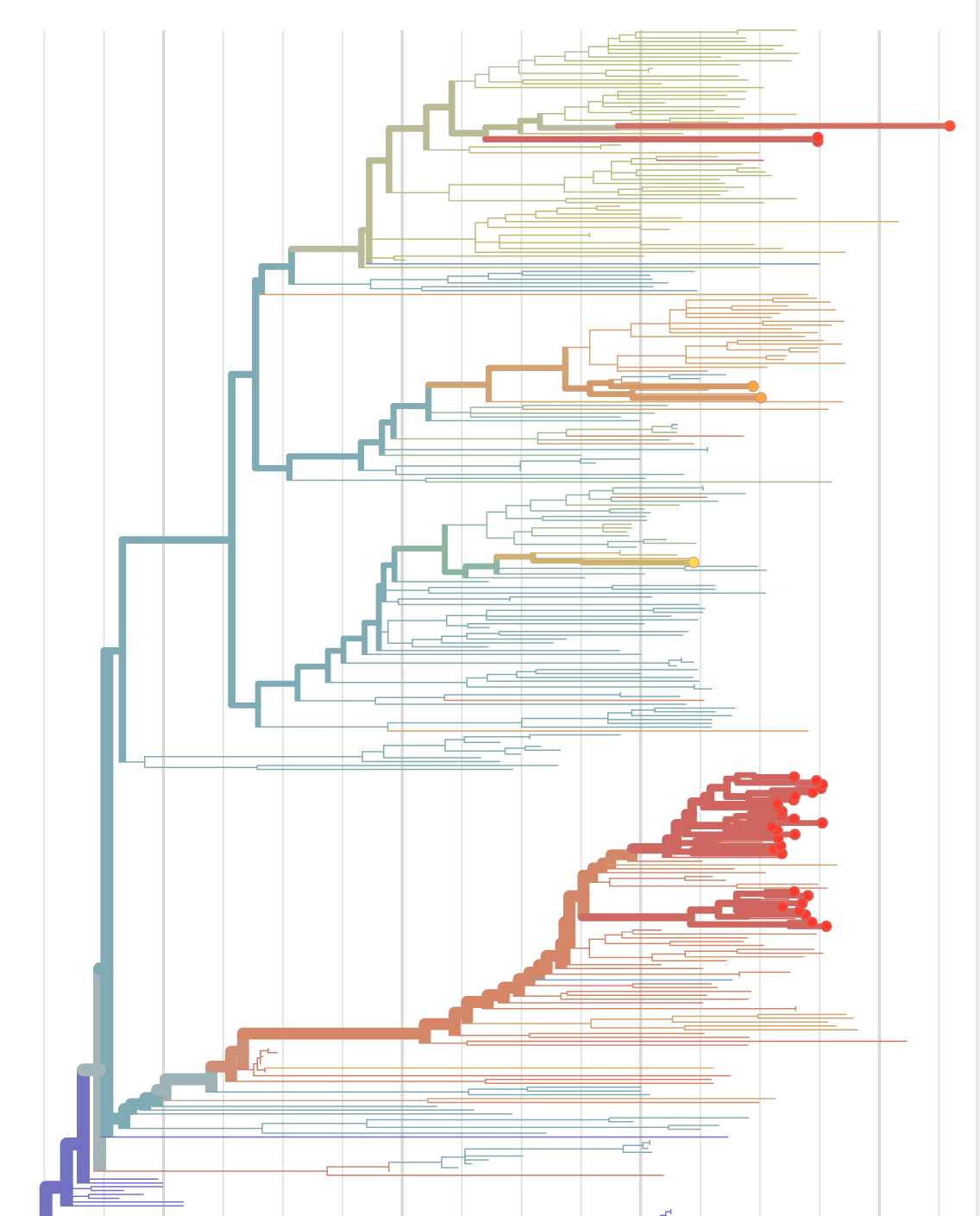
Sequences record the spread of pathogens




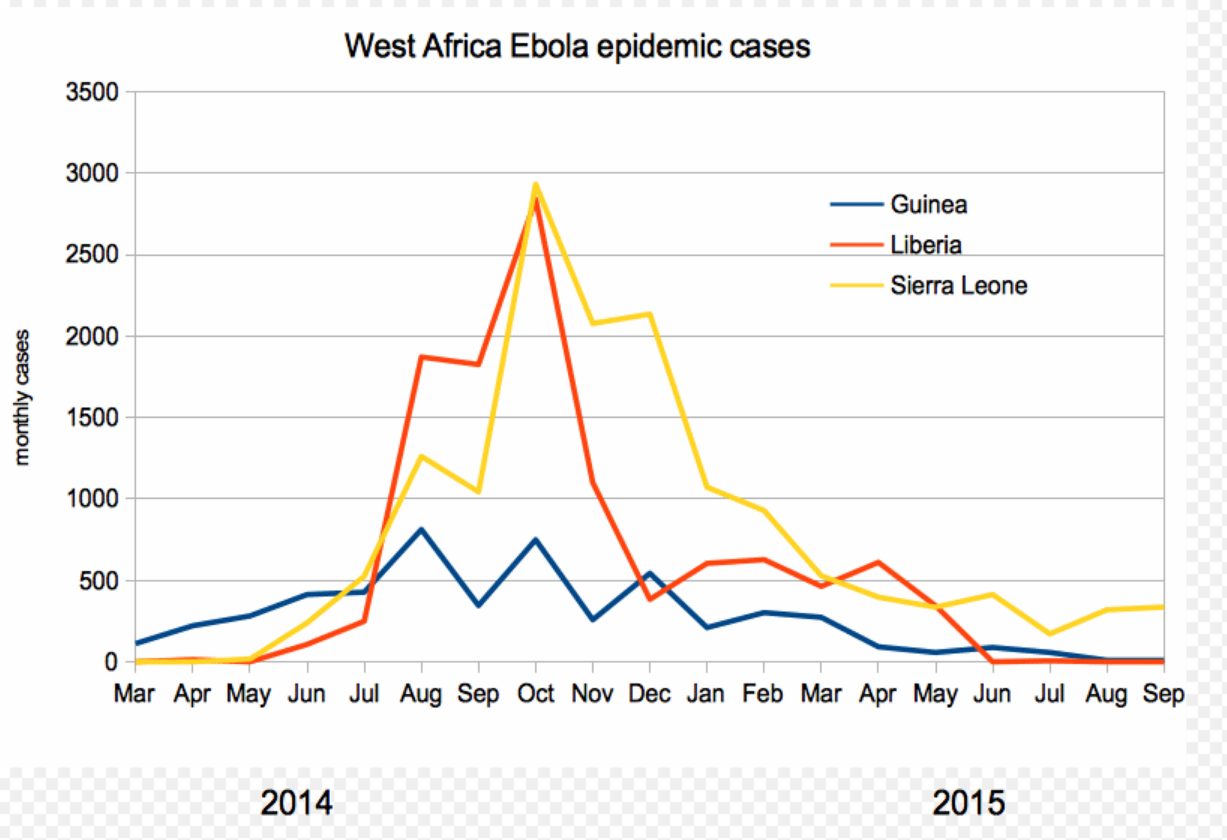
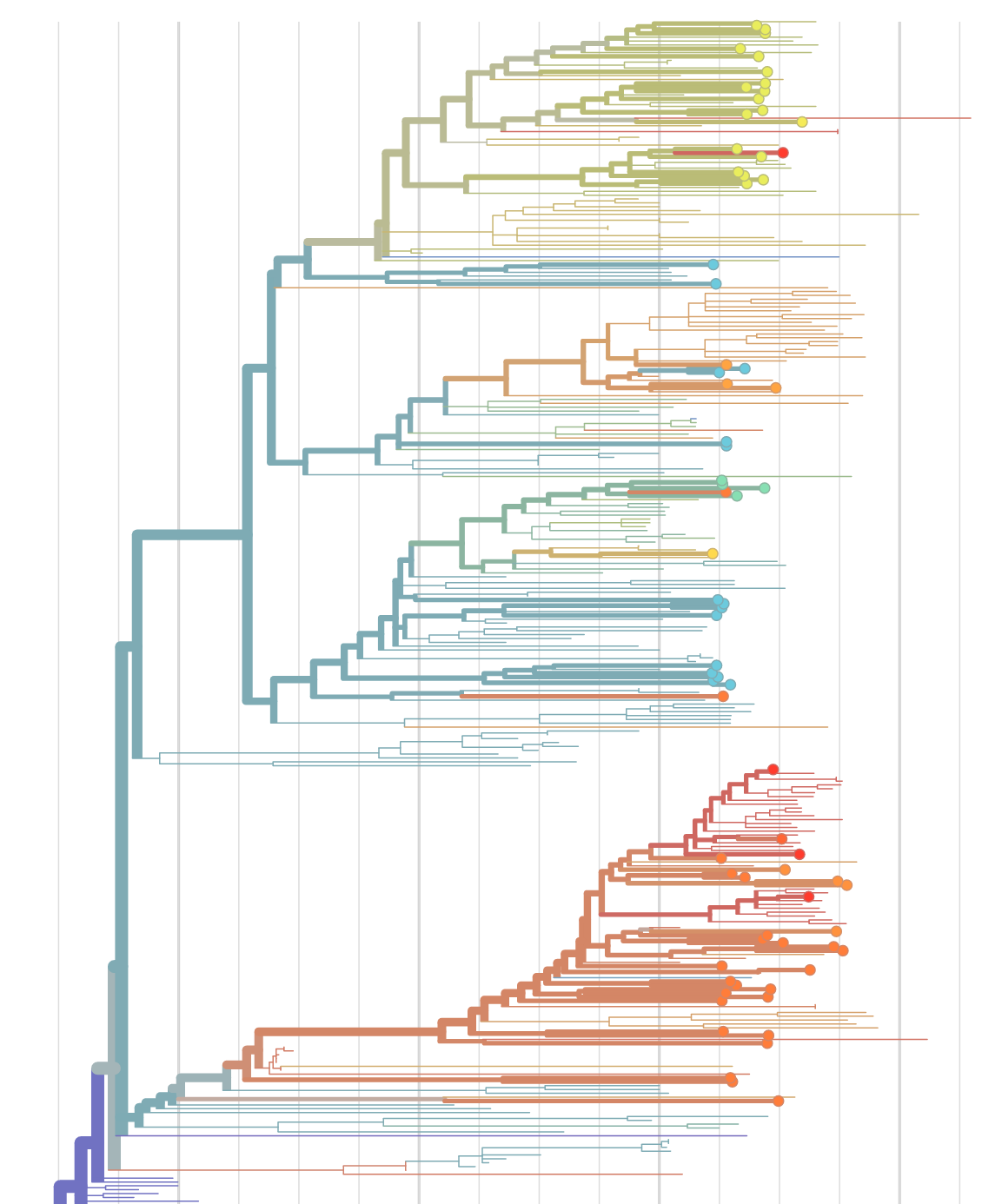
2013-2015 West African Ebola virus outbreak
It took two years to have a detailed understanding of the outbreak. Why did this take so long?
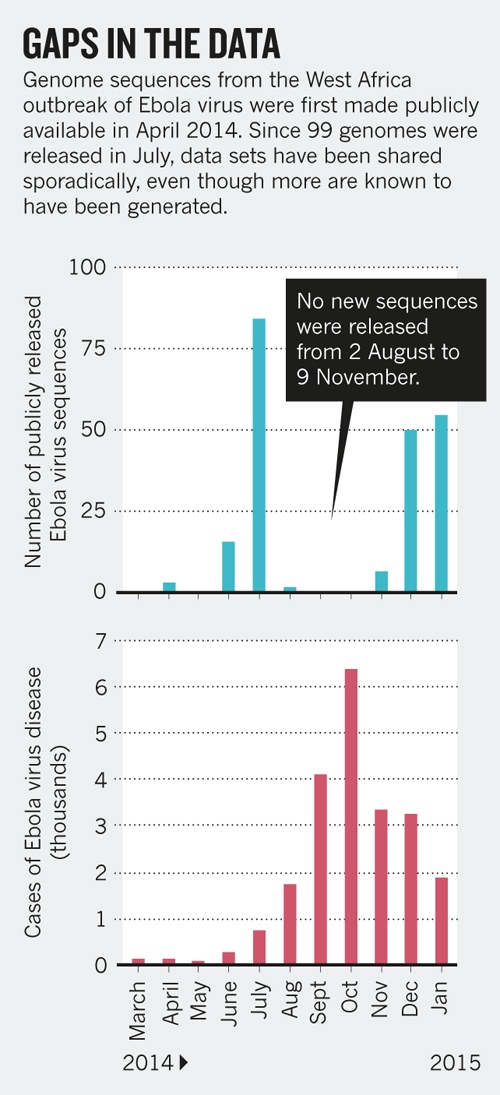
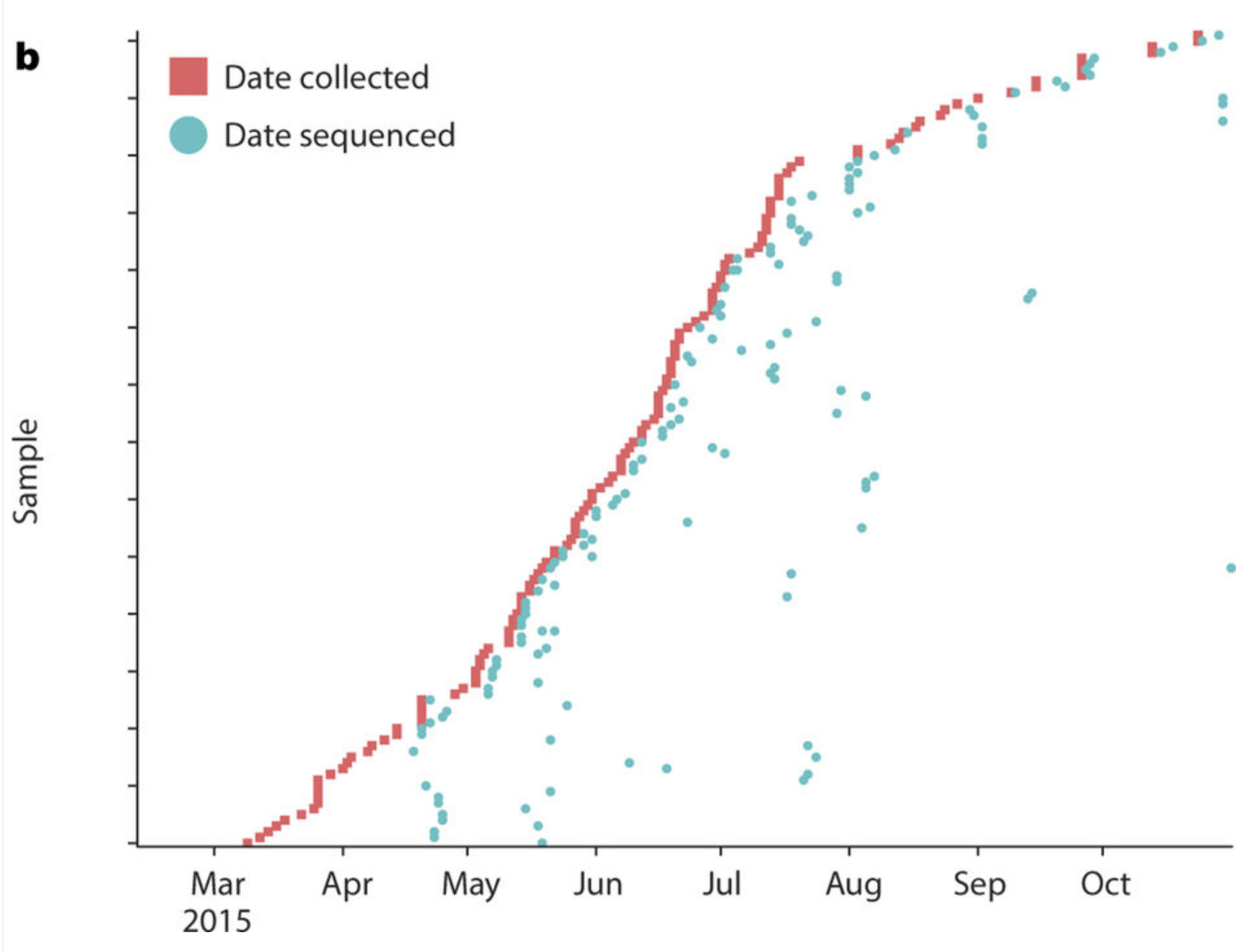
Sharing of Ebola virus sequences
- Baize et al.: Samples collected in March 2014, sequences in GenBank within a month
- Gire et al.: released data as it was generated
- Followed by a long gap...
- Early insights into transmission dynamics are important
- Sharing has to be immediate -- not upon publication
Recurring problem
- 2002 Severe acute respiratory syndrome (SARS)
- 2003 H5N1 influenza outbreak. Some countries stopped sharing any data
- 2013-2015 Ebola virus outbreak in West Africa
- 2014-2016 Zika Virus outbreak: Controversies about attribution and reuse
- 2014- H7N9 influenza outbreak: Controversies about attribution and reuse
Different disease -- different scientists and institutions.
→Lessons need to be relearned.
Barriers to data sharing: scientists
- Privacy of study participants
- Fear of being scooped/ensure maximal return
- Secondary analysis perceived as freeloading: "data parasites"
- Don't want to be second guessed
- Release and curation is laborious
- Sloppy records
Also see Smith et al, F1000 2016
Barriers to data sharing: organizations and governments
- Economic consequences of outbreaks (tourism, agriculture)
- Conflicts between high and low/middle income countries
- Concerns about IP and commercial exploitation
- Legislative/regulatory barriers
See also: van Panhius et al, BMC Public health 2014
Overcoming Barriers
Open vs restricted sharing
- Alternatives to GenBank: GenBank is "public domain", no requirement to credit data producers
- GISAID/EpiFlu: sign-up and agree to terms and conditions

- virological.org: Platform for sharing and discussing molecular epidemiology
- Explicit data reuse terms
- Outline planned projects in white-paper
- Caveat: Very difficult to enforce...
See Bogner et al, 2006
Building Trust
- Peter Bogner coordinated Influenza data sharing
- Andrew Rambaut coordinated Ebola virus data sharing
- During the EBV outbreak, WHO and journals explicitly encouraged data sharing
See also Smith et al, F1000 2016
Make sharing easy and provide incentives!
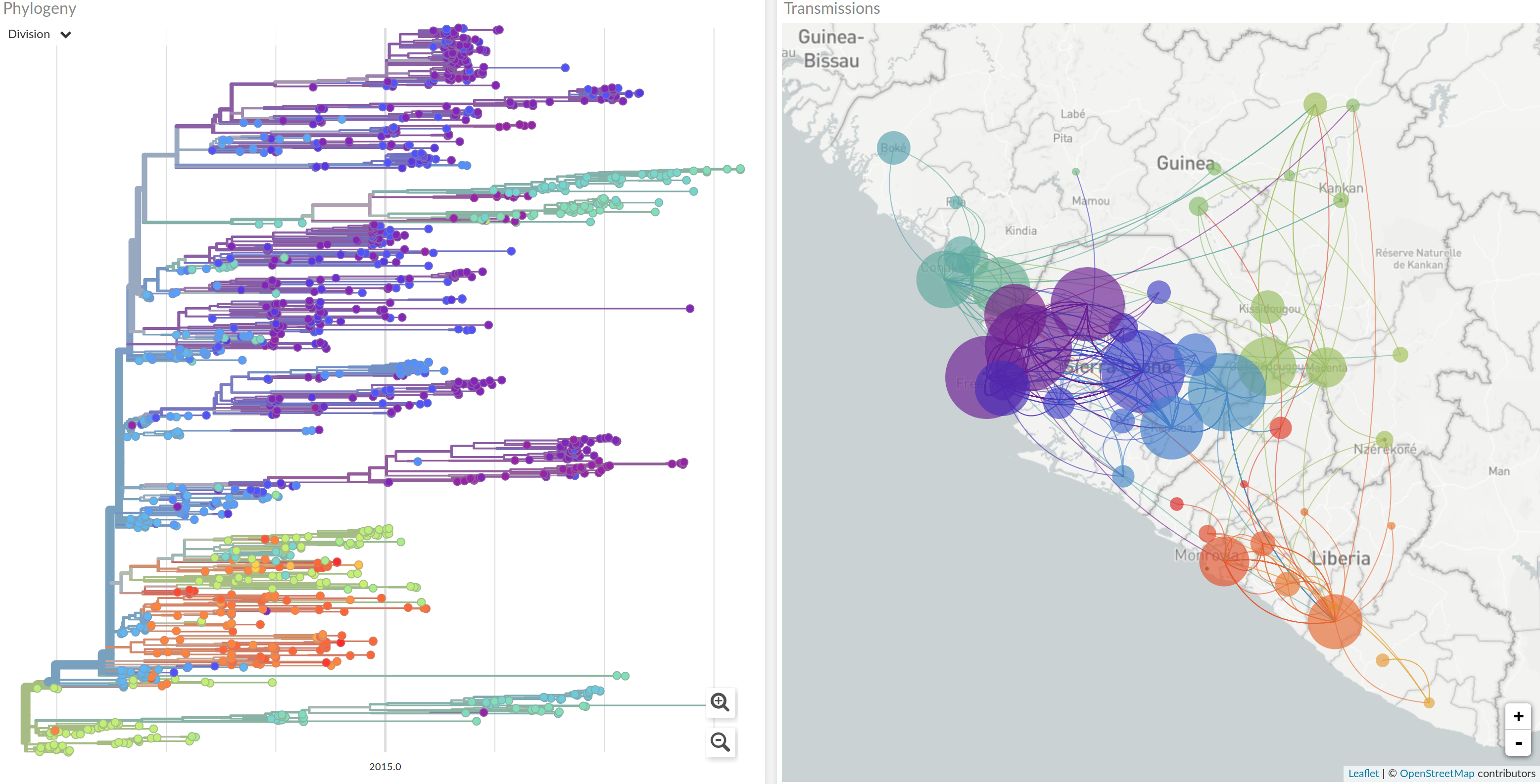
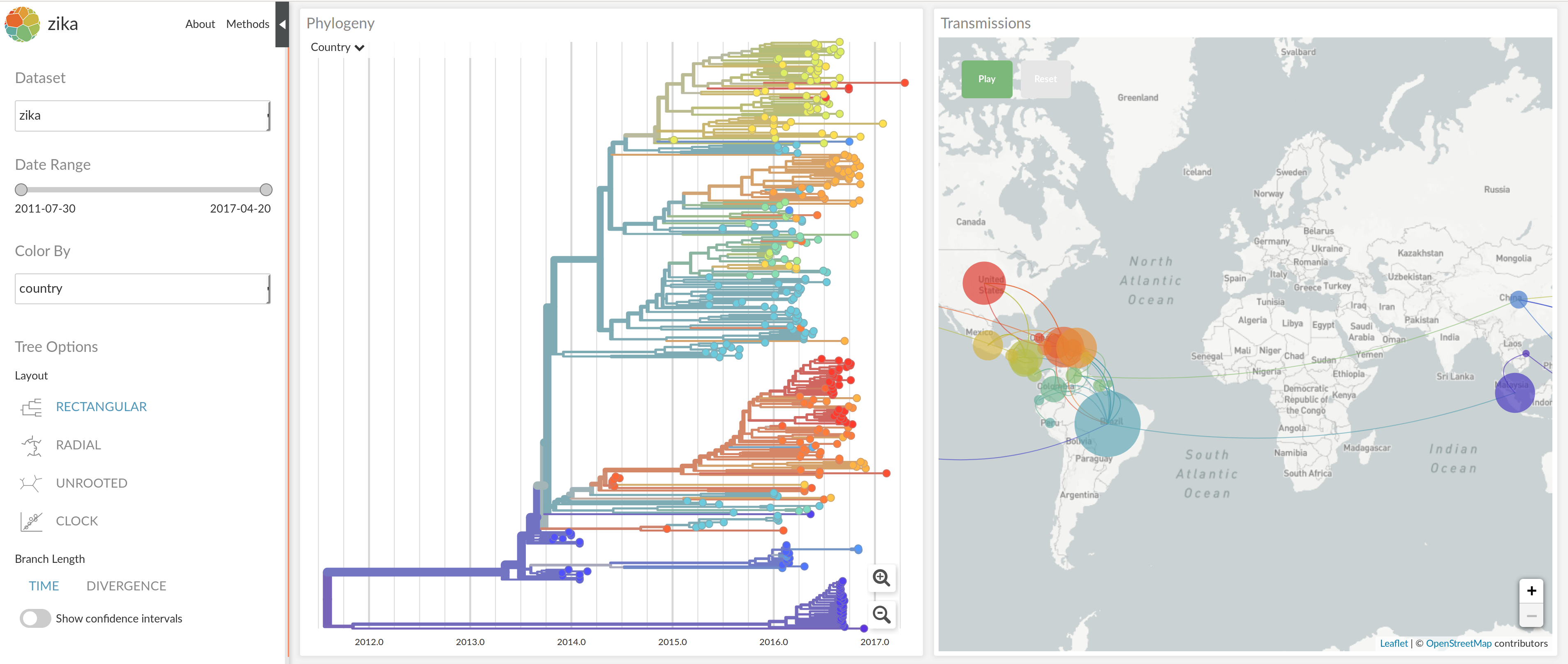
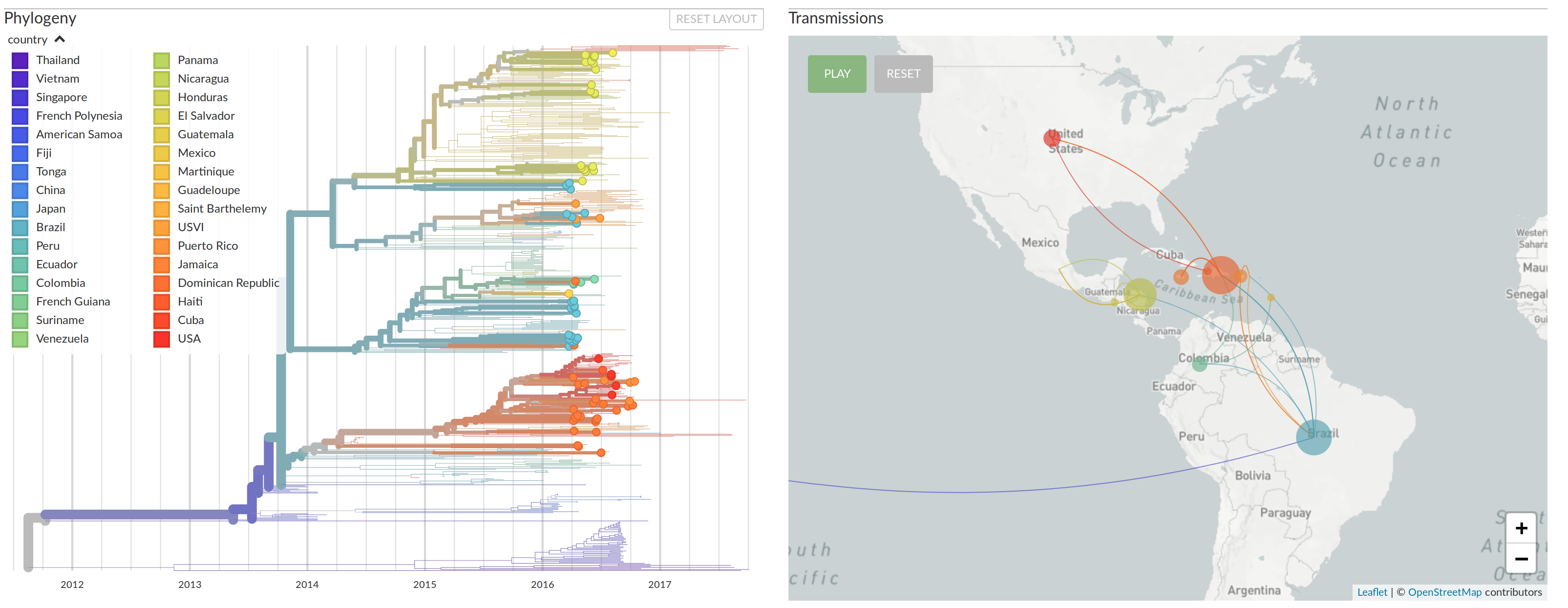
nextstrain.org
joint work with Trevor Bedford & his lab
code at github.com/nextstrain
NextStrain architecture
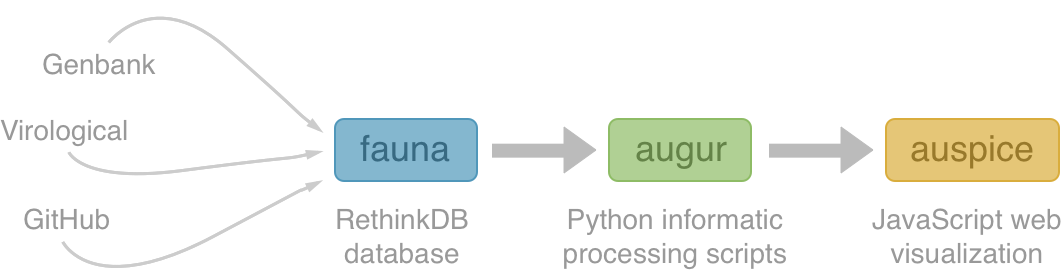
Using treetime to rapidly compute timetrees
Real-time influenza virus analysis with nextstrain
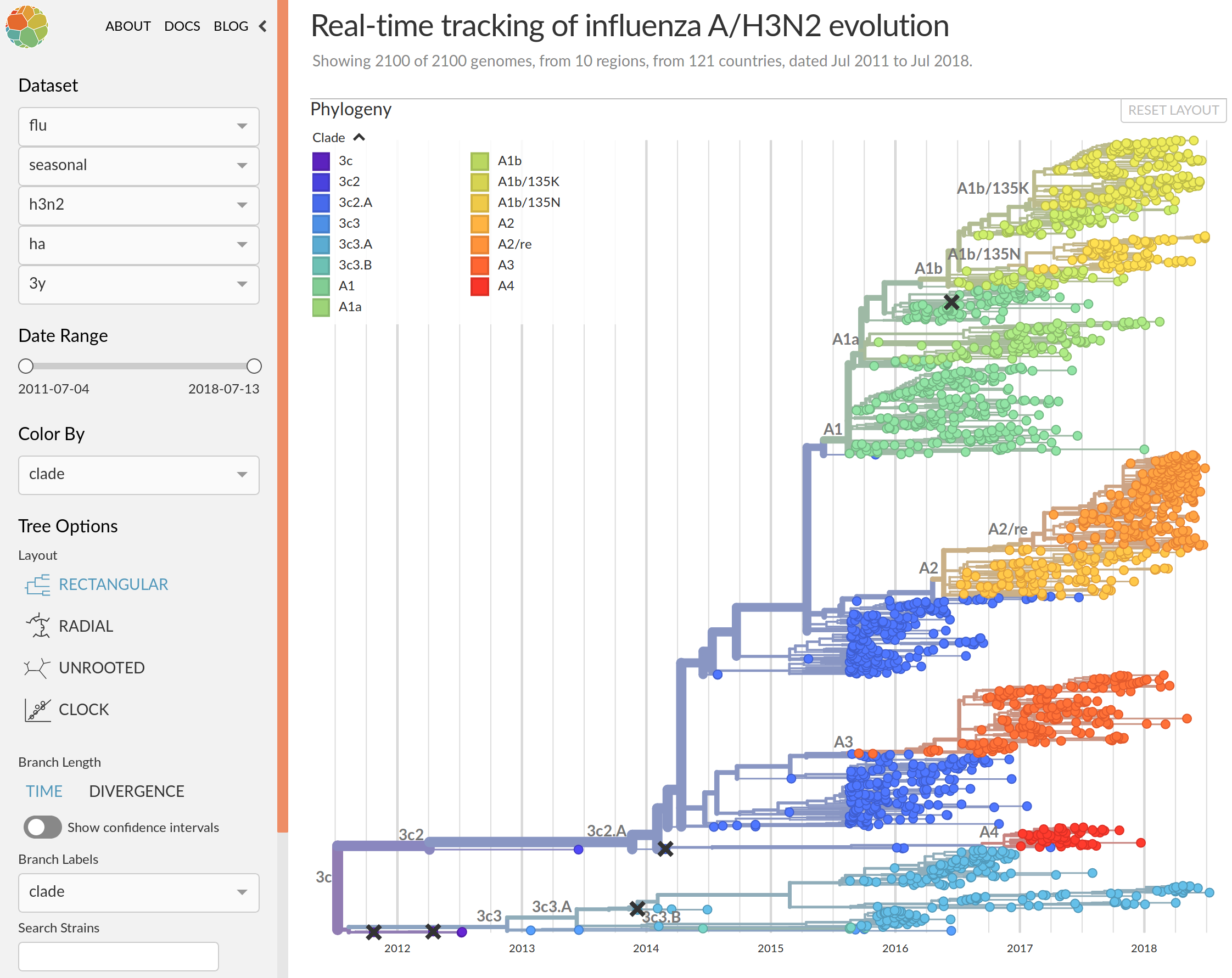
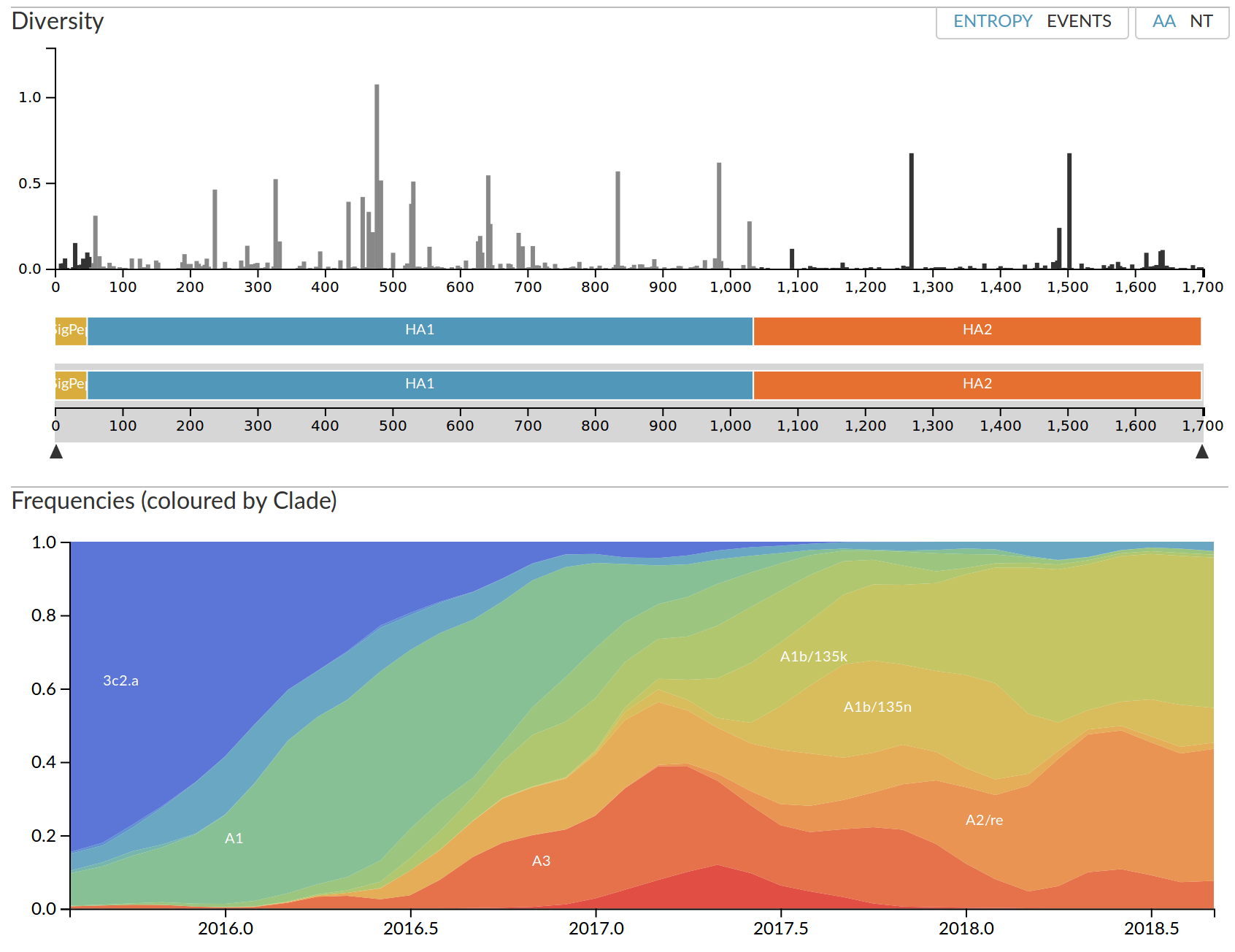
nextstrain.org
- Global analysis provides context for new sequences
- Once nextstrain became the largest collection of Ebola/Zika sequences, everybody wanted their sequences on nextstrain
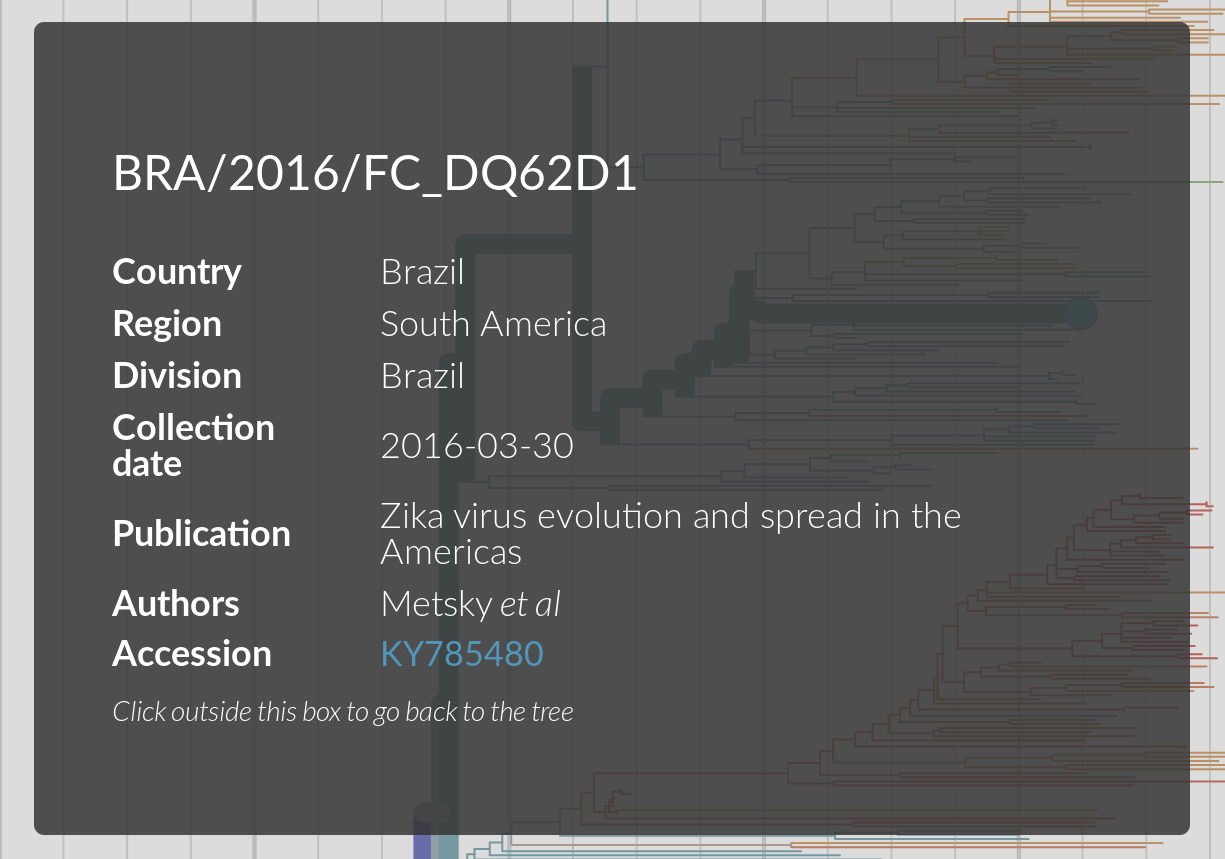
- We take care to highlight the scientists contributing data
- Link to original source, rather than reshare
- nextstrain.org/zika?f_authors=Metsky et al
Open source: github.com/nextstrain
Summary and outlook
- In public health emergencies, immediate data sharing is imperative
- Response has to be fast
→ pre-existing trusted framework to share data
→ rapid and readily deployable analysis tools - We need incentives for high quality data sets
- We need better ways to credit data producers
→ data citations - Wider acceptance of preprints should help
→ establishes priority and a citation hook
nextstrain.org
- Trevor Bedford
- Colin Megill
- Pavel Sagulenko
- Sidney Bell
- James Hadfield
- Wei Ding
- Tom Sibley
- Emma Hodcroft



Data symbiont award