Outlook: Surveillance with Nextstrain
Richard Neher
Biozentrum, University of Basel
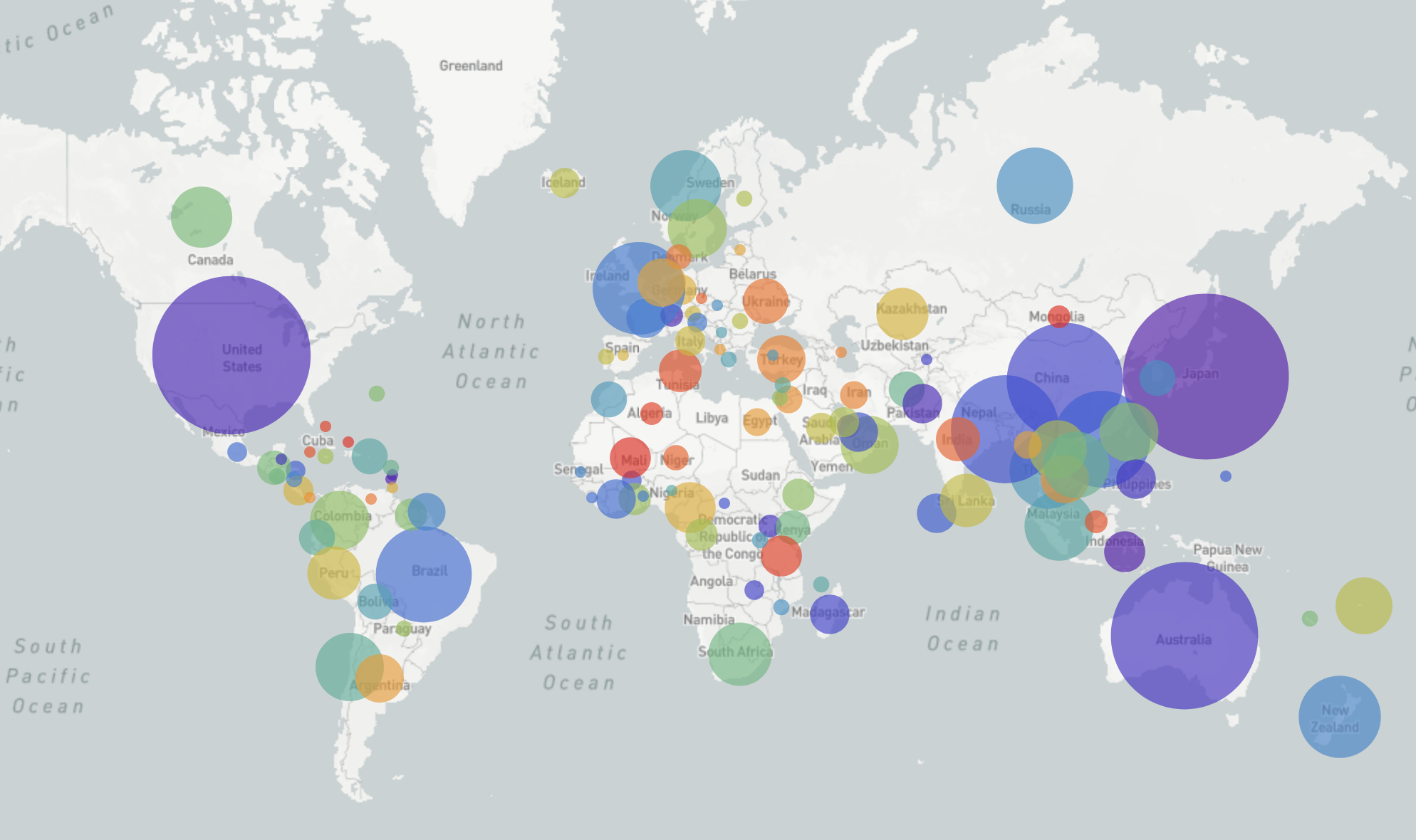
GISRS and GISAID -- Influenza virus surveillance
- comprehensive coverage of the world
- timely sharing of data -- often within 2-3weeks of sampling
- hundreds of sequences per week (in peak months)
→ requires continuous analysis and easy dissemination
→ interpretable and intuitive visualization
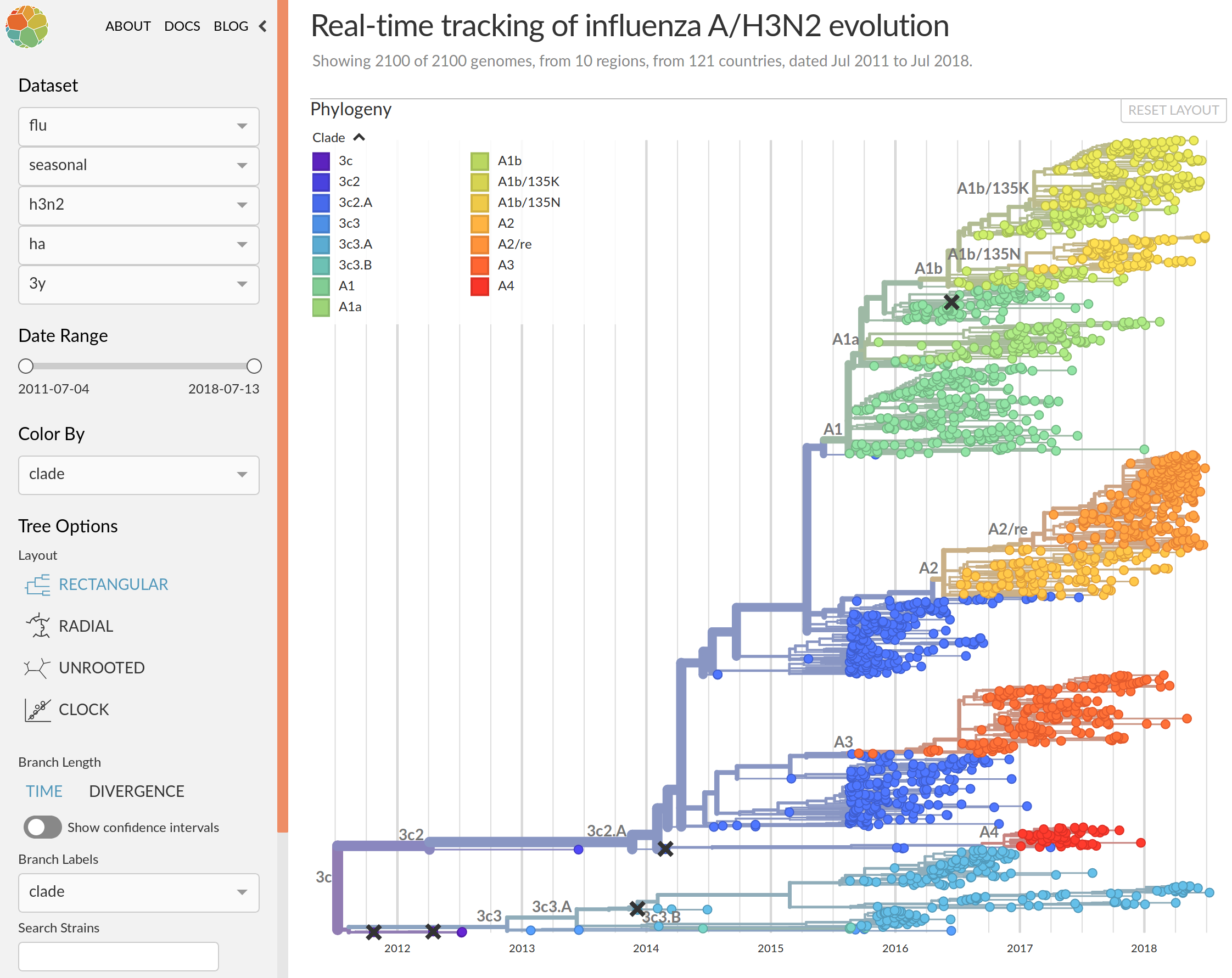
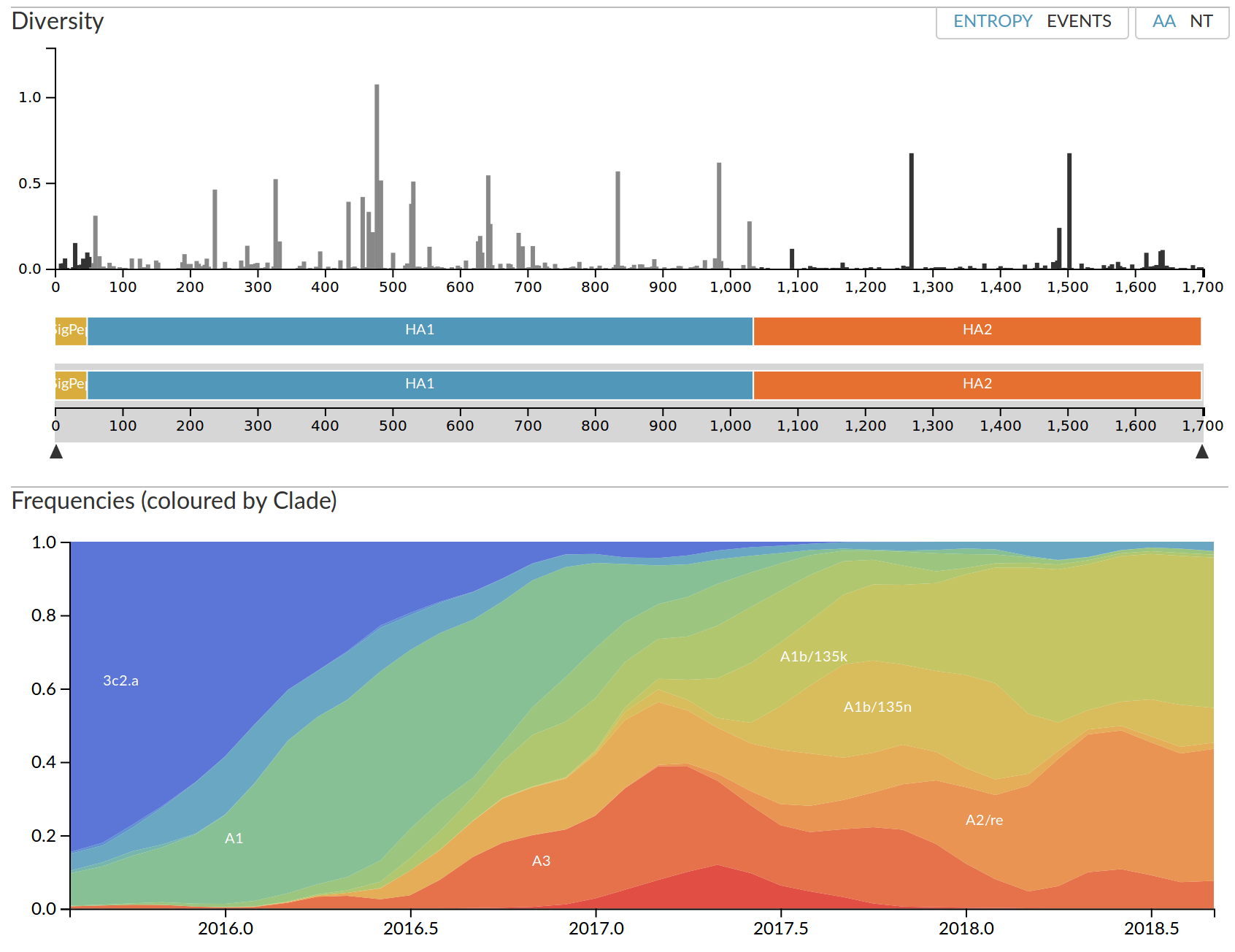
Real-time analysis with nextstrain
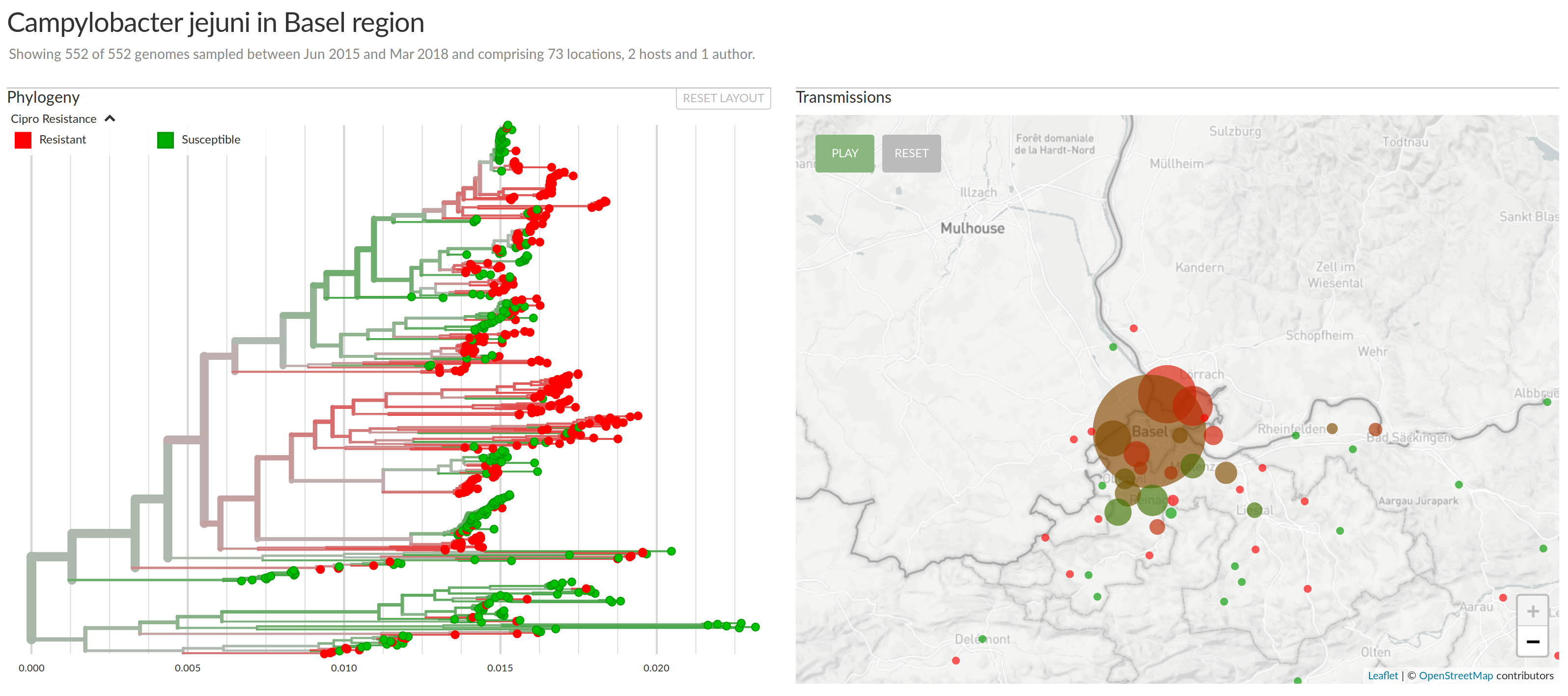
Campylobacter Jejeuni in the Basel area (Reist, Seth-Smith et al)
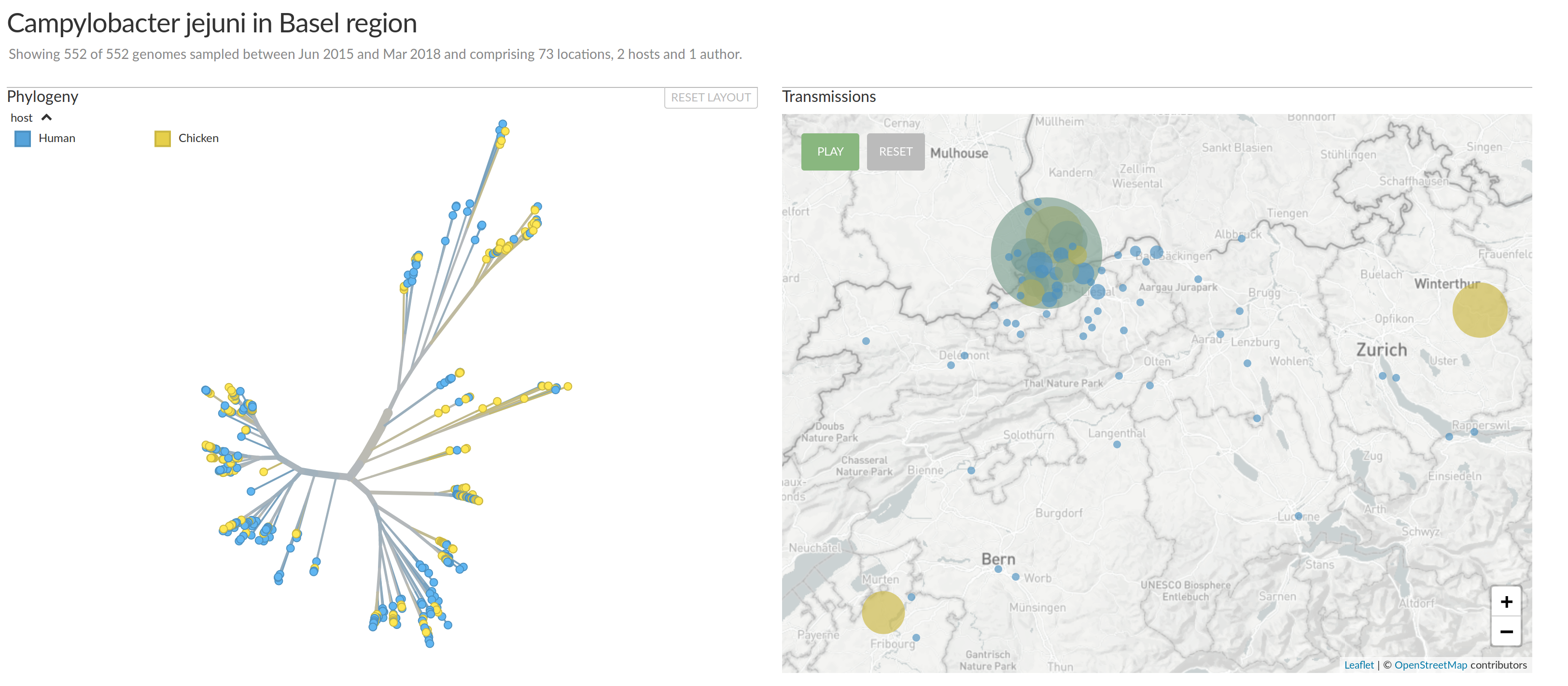
Campylobacter Jejeuni in the Basel area (Reist, Seth-Smith et al)
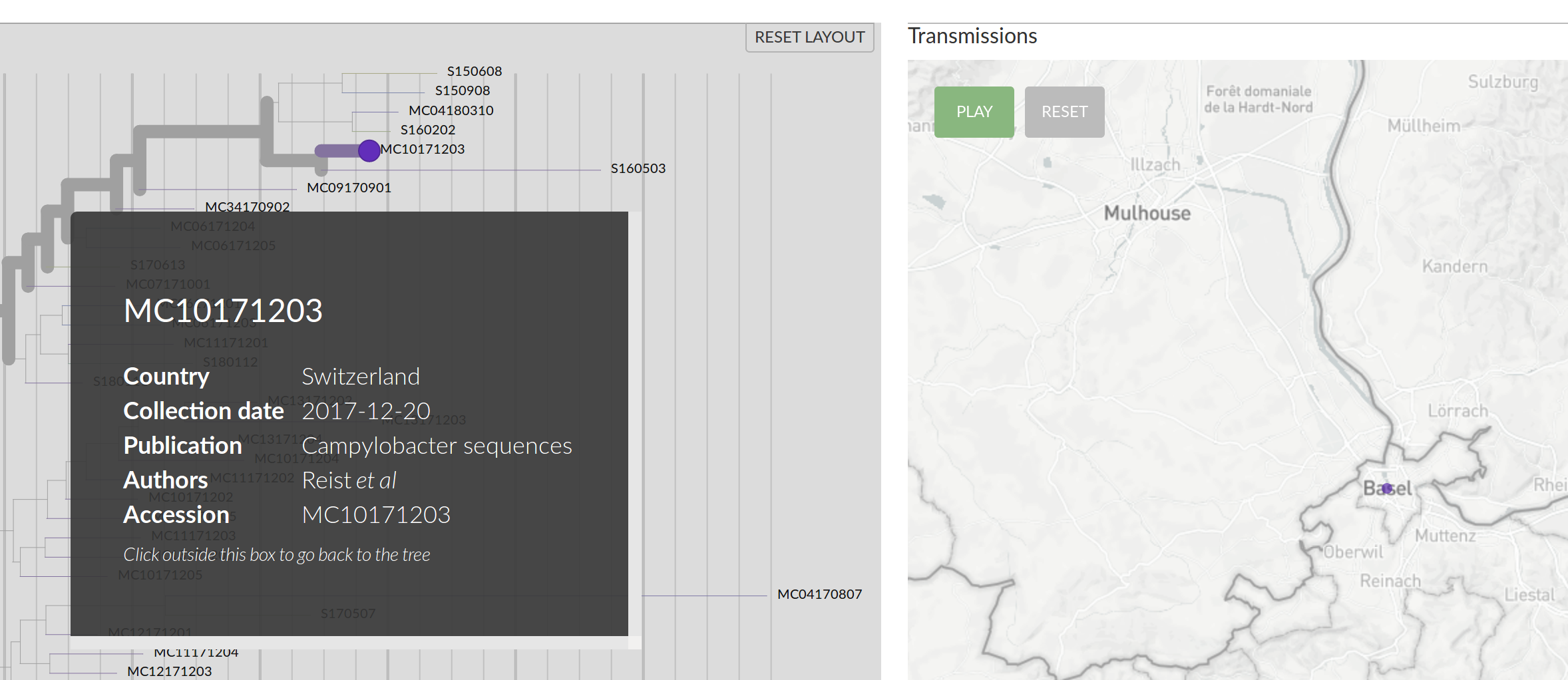
Ciprofloxacin resistance (Reist, Seth-Smith et al)
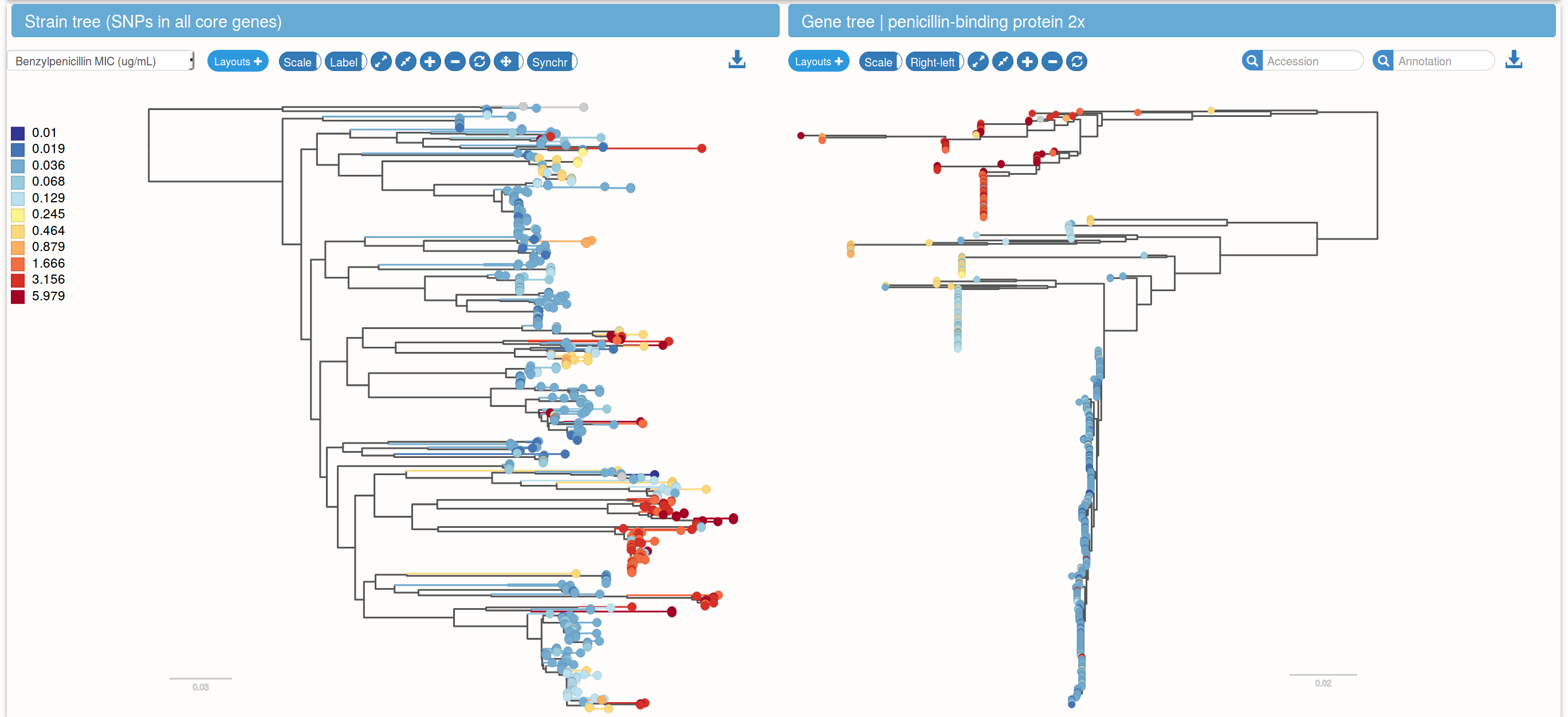
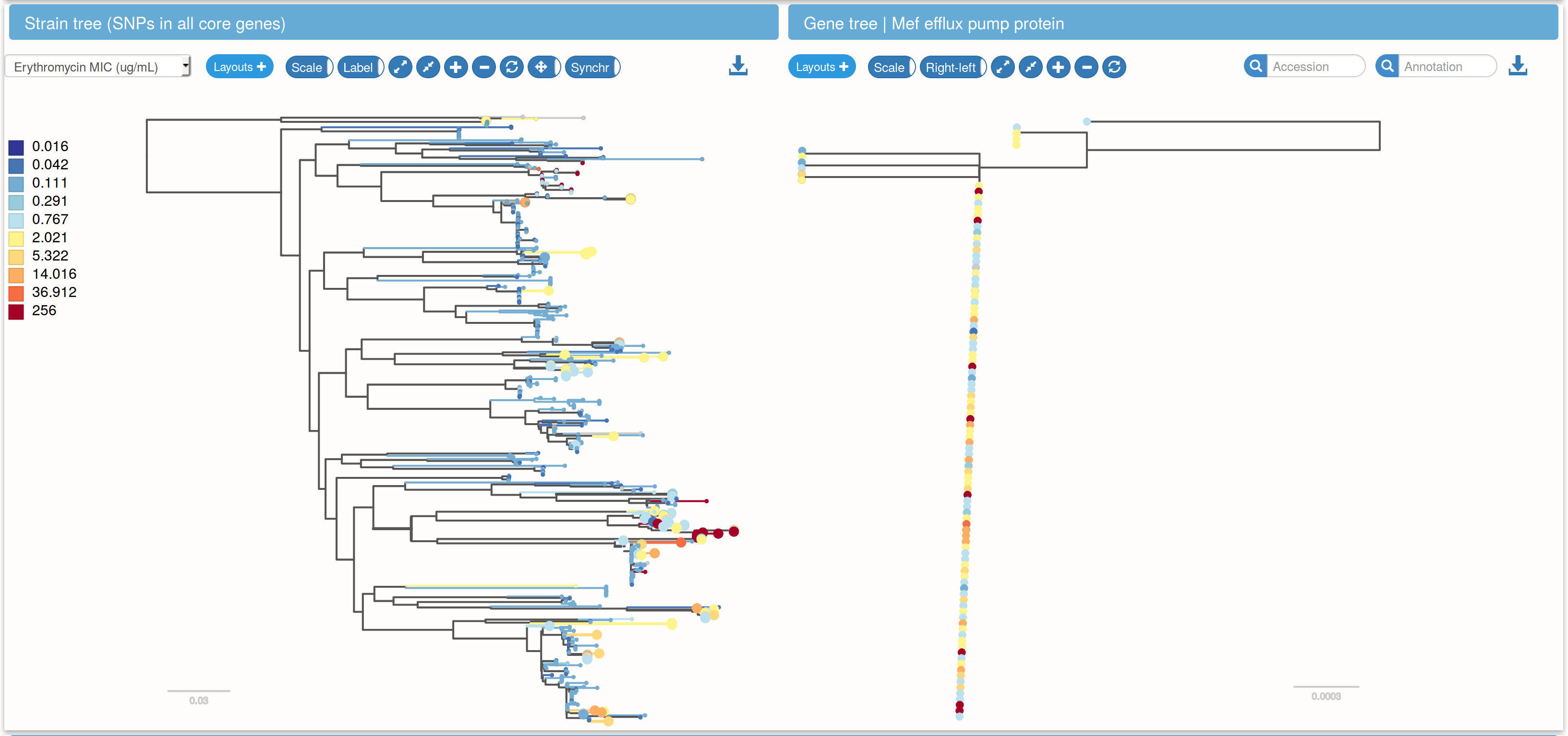
Species trees and gene trees
Phenotype associations with acquired genes
Plasmid phylogenies

Enterovirus D68 -- Collaboration with Jan Albert and Robert Dyrdak
- Non-polio enterovirus
- Large outbreak in 2014 with severe neurological symptoms in young children (acute flaccid myelitis)
- Another outbreak in 2016
- Outbreaks tend to start in late summer/fall
- Several reports of EV-D68 outbreaks in the past 6 weeks
(127 AFM cases in the US as of yesterday)
Whole genome deep sequencing
- Geographic spread and phylogenetic patterns?
- Immune escape?
- Within host diversity?
- Transmission bottlenecks/multiplicity of infection?
nextstrain.org/enterovirus
joint work with Jan Albert & his lab
Phylodynamic analysis
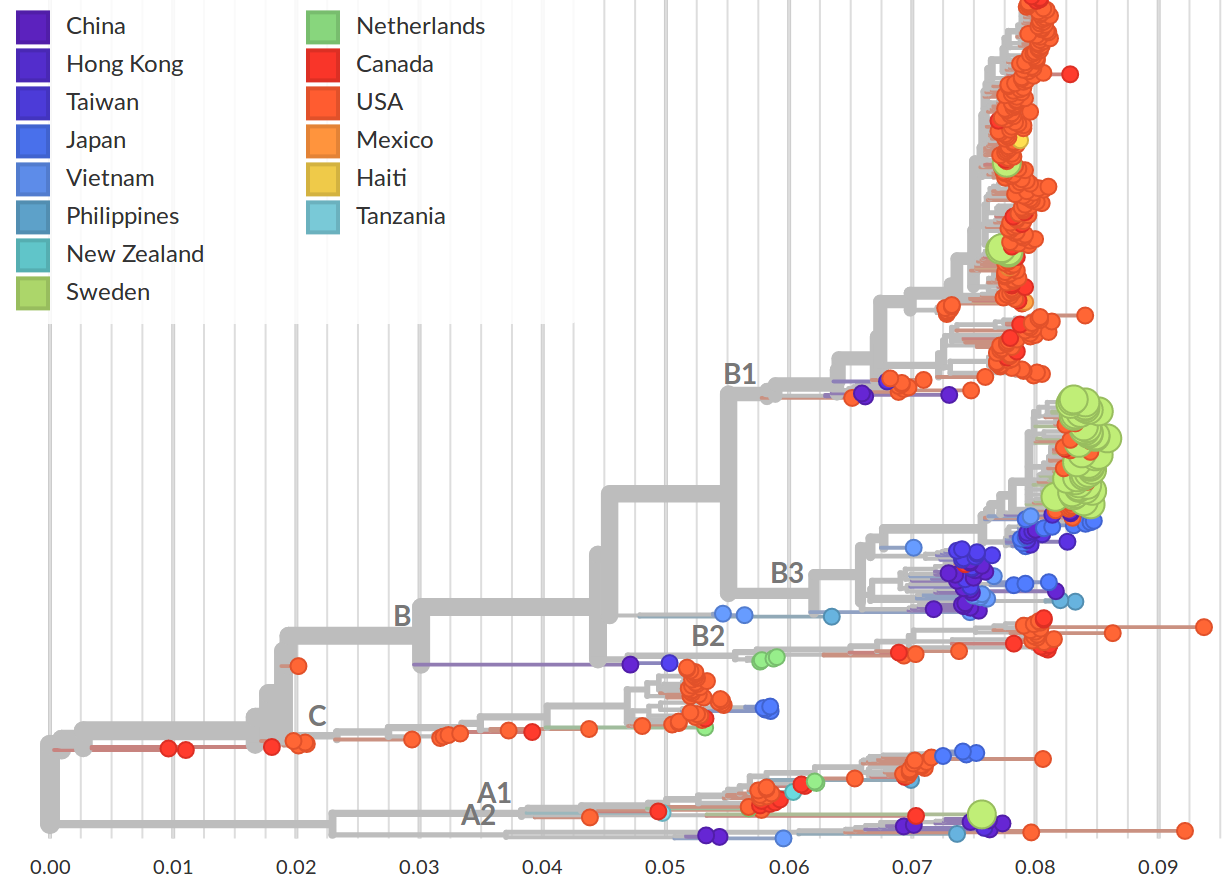
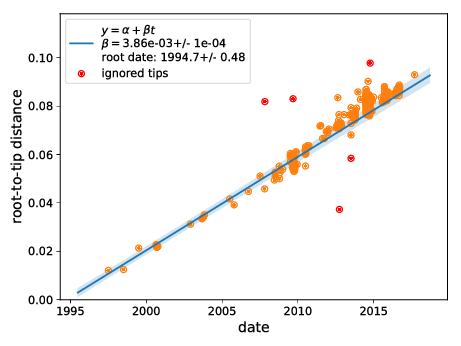
- EV-D68 outbreaks come from distinct clades.
- The evolutionary rate is very high -- a lot of power to study transmission chains
- Most variation is synonymous
Acknowledgments






- Trevor Bedford
- Colin Megill
- Pavel Sagulenko
- Sidney Bell
- James Hadfield
- Wei Ding
- Emma Hodcroft
- Sanda Dejanic




Acknowledgments
- Robert Dyrdak
- Jan Albert
- Lina Thebo
- Emma Hodcroft
