Virus evolution and the spread of infectious disease
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/201812_Virology.html
Human Influenza A viruses

Weekly numbers of positive influenza tests in the US by subtype
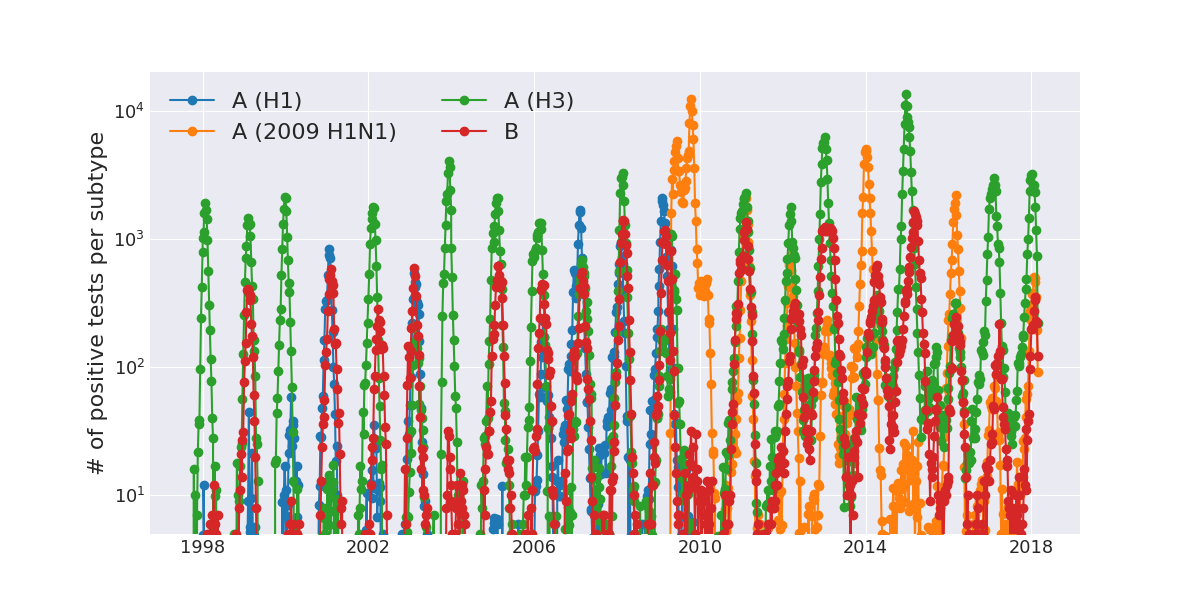 Data by the US CDC
Data by the US CDC
Influenza virus
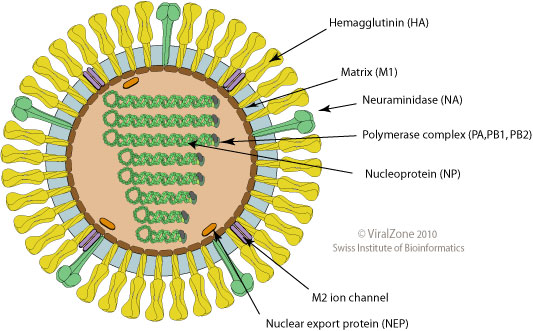
- Surface proteins hemagglutinin (HA) and neuraminidase (NA)
- Influenza A virus
- Common in birds and mammals
- Many different subtypes defined by surface proteins
- H3N2, H1N1, H7N9, H5N1
- Influenza B virus
- infects mainly humans
- two lineages that split 30-40y ago
- B/Victoria vs B/Yamagata



- Influenza virus evolves to avoid human immunity
- Vaccines need frequent updates
Vaccine selection time line
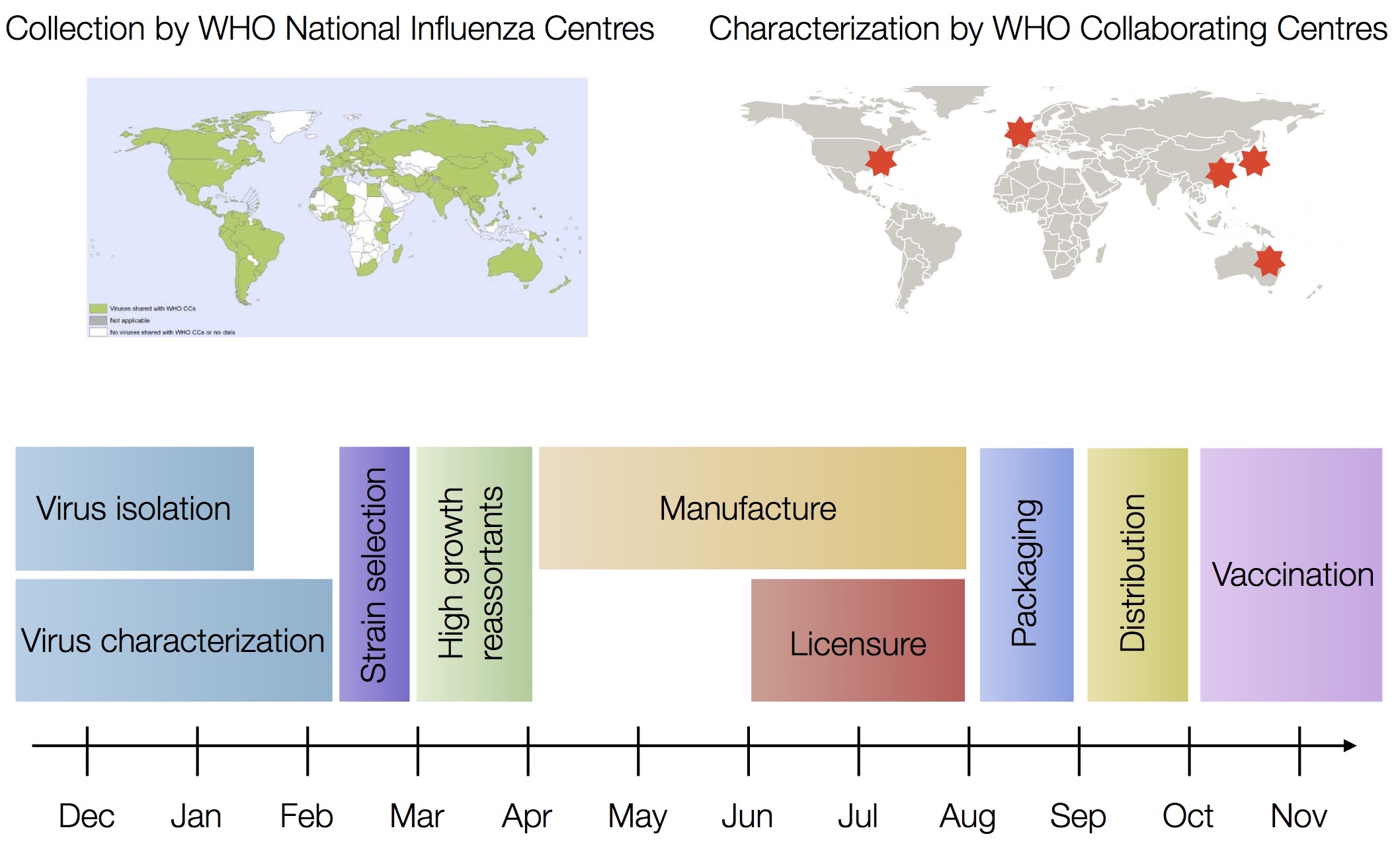
Slide by Trevor Bedford
Tracking virus spread and evolution by sequencing
A/Brisbane/100/2014
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
A/Brisbane/1000/2015
GGATAATTCTATTAACCATGAAGACTATTATTGCTTT...
A/Brisbane/1/2017
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
... thousands of sequences...
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
A/Brisbane/1000/2015
GGATAATTCTATTAACCATGAAGACTATTATTGCTTT...
A/Brisbane/1/2017
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
... thousands of sequences...
Phylogenetic analysis of viral sequences
RNA viruses have a high mutation rate. New mutations arise every few weeks.
nextflu.org
joint work with Trevor Bedford & his lab
Hemagglutination Inhibition assays
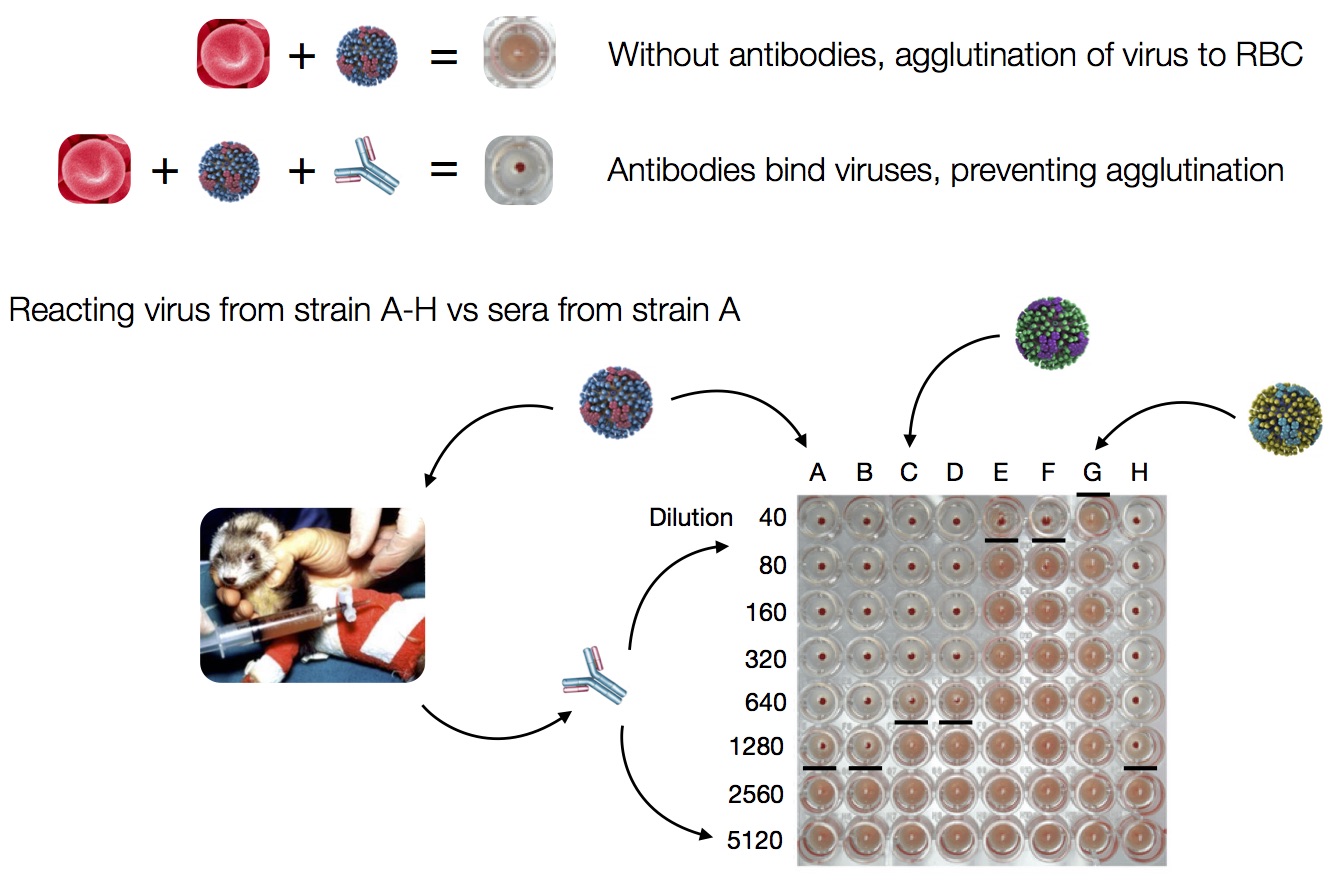
HI data sets
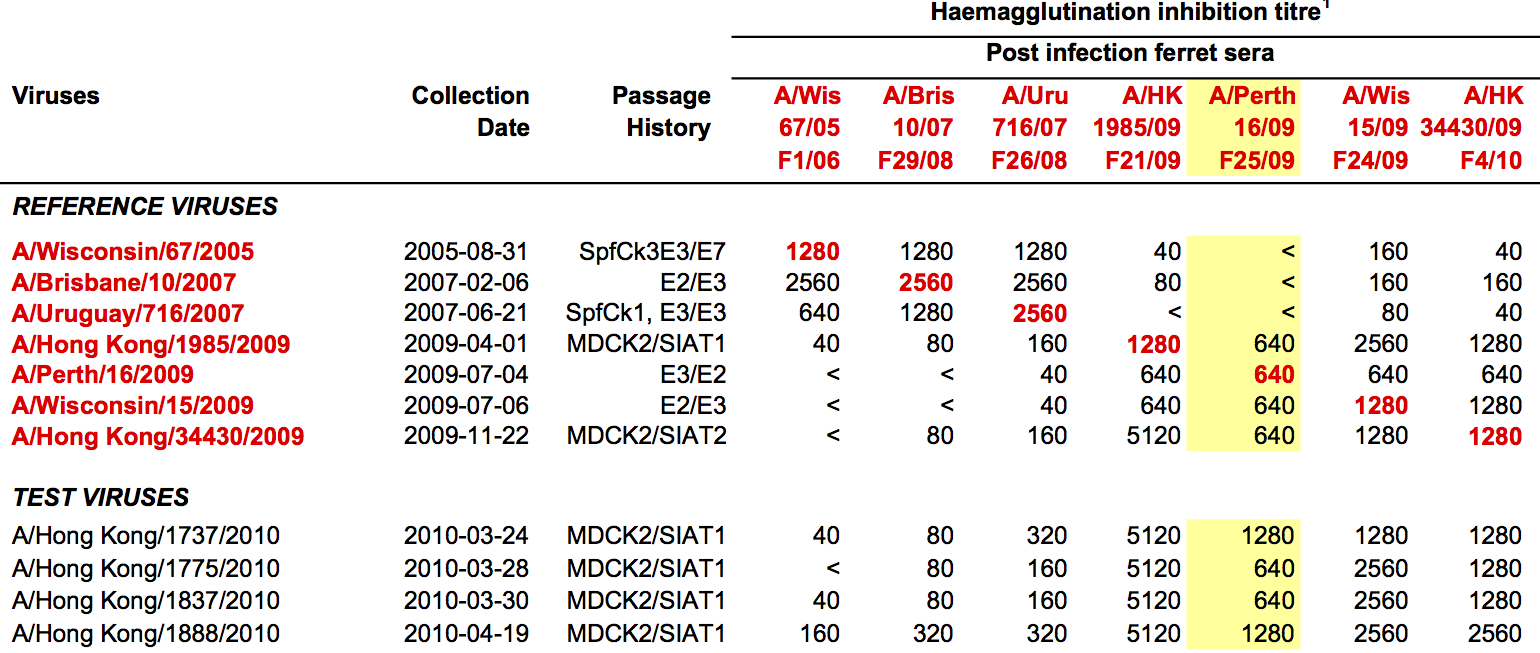
- Long list of distances between sera and viruses
- Tables are sparse, only close by pairs
- Structure of space is not immediately clear
- MDS in 2 or 3 dimensions
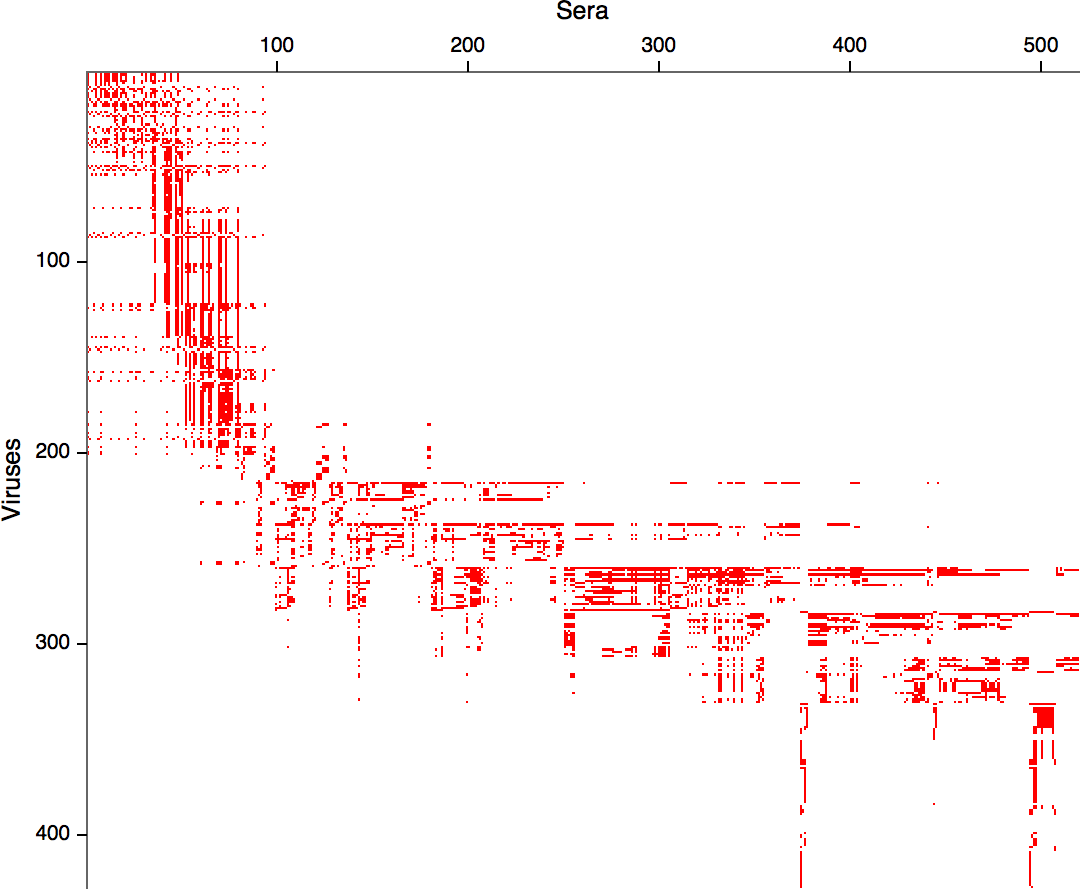
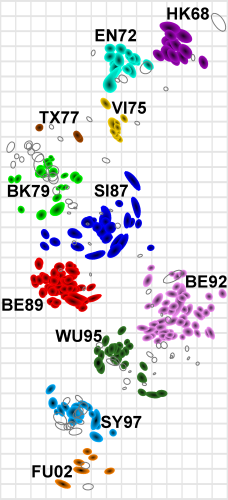 Smith et al, Science 2002
Smith et al, Science 2002
Antigenic evolution
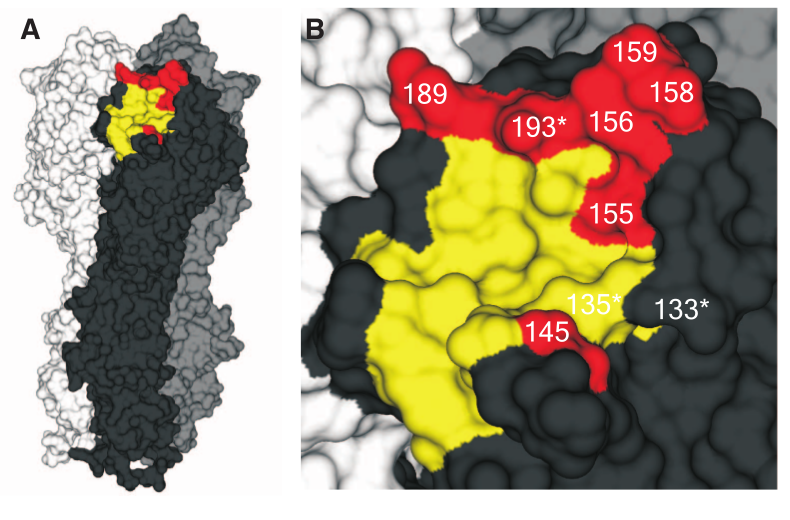

Evolution of HIV
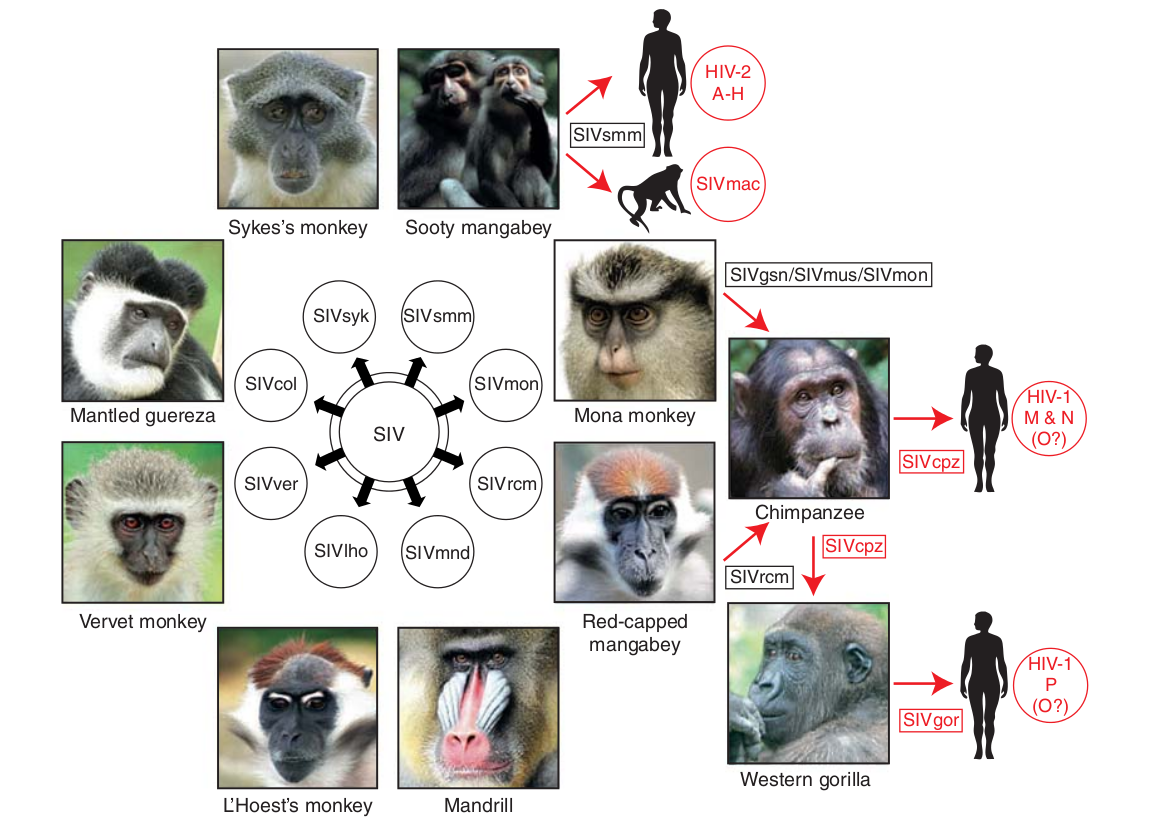
- Chimp → human transmission around 1900 gave rise to HIV-1 group M
- ~100 million infected people since
- subtypes differ at 10-20% of their genome
- HIV-1 evolves ~0.1% per year
HIV-SIV phylogeny
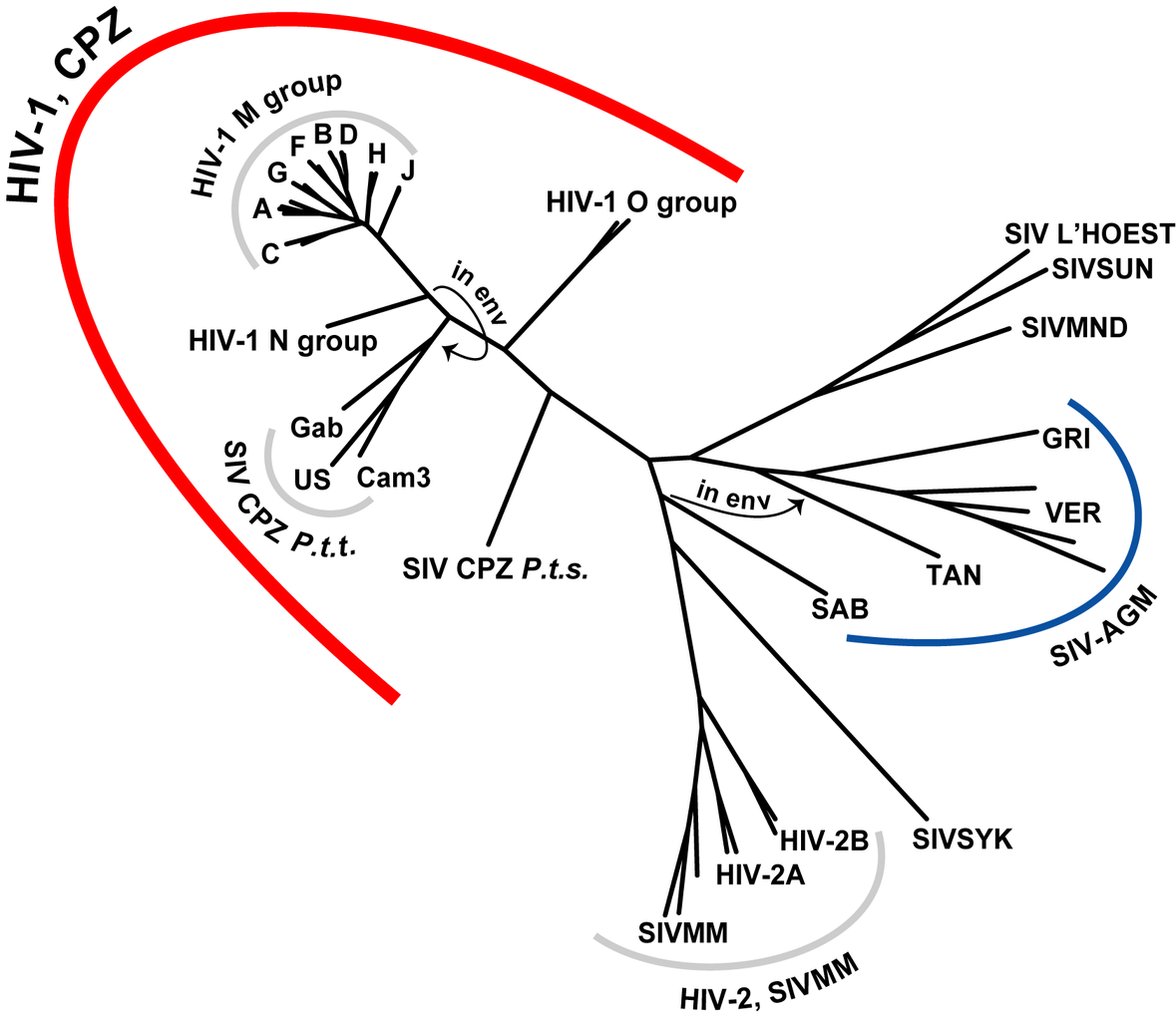 wikipedia.org
wikipedia.org
What makes these viruses so good at evolving?
High mutation rates
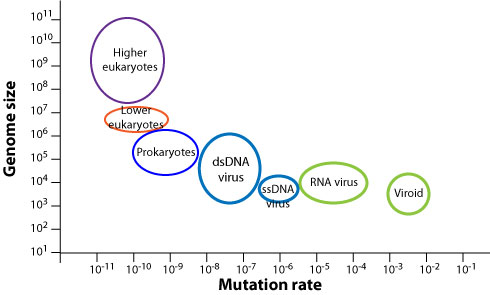
viralzone.expasy.org
Large population sizes
- Within a chronically infected person, HIV infects $10^8$ cells a day
- Influenza viruses infects $10^8$ humans a year
- every infected cell produces 1000s of virions
- → every possible mutation is produced many times every day
Recombination
Influenza Virus
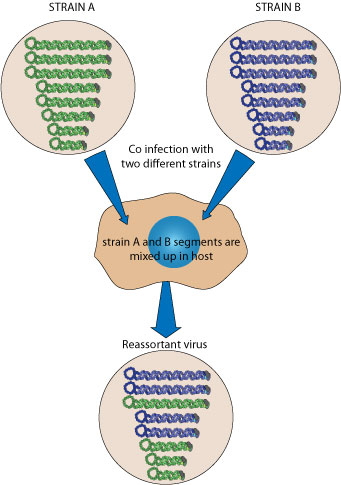
viralzone.expasy.org
HIV
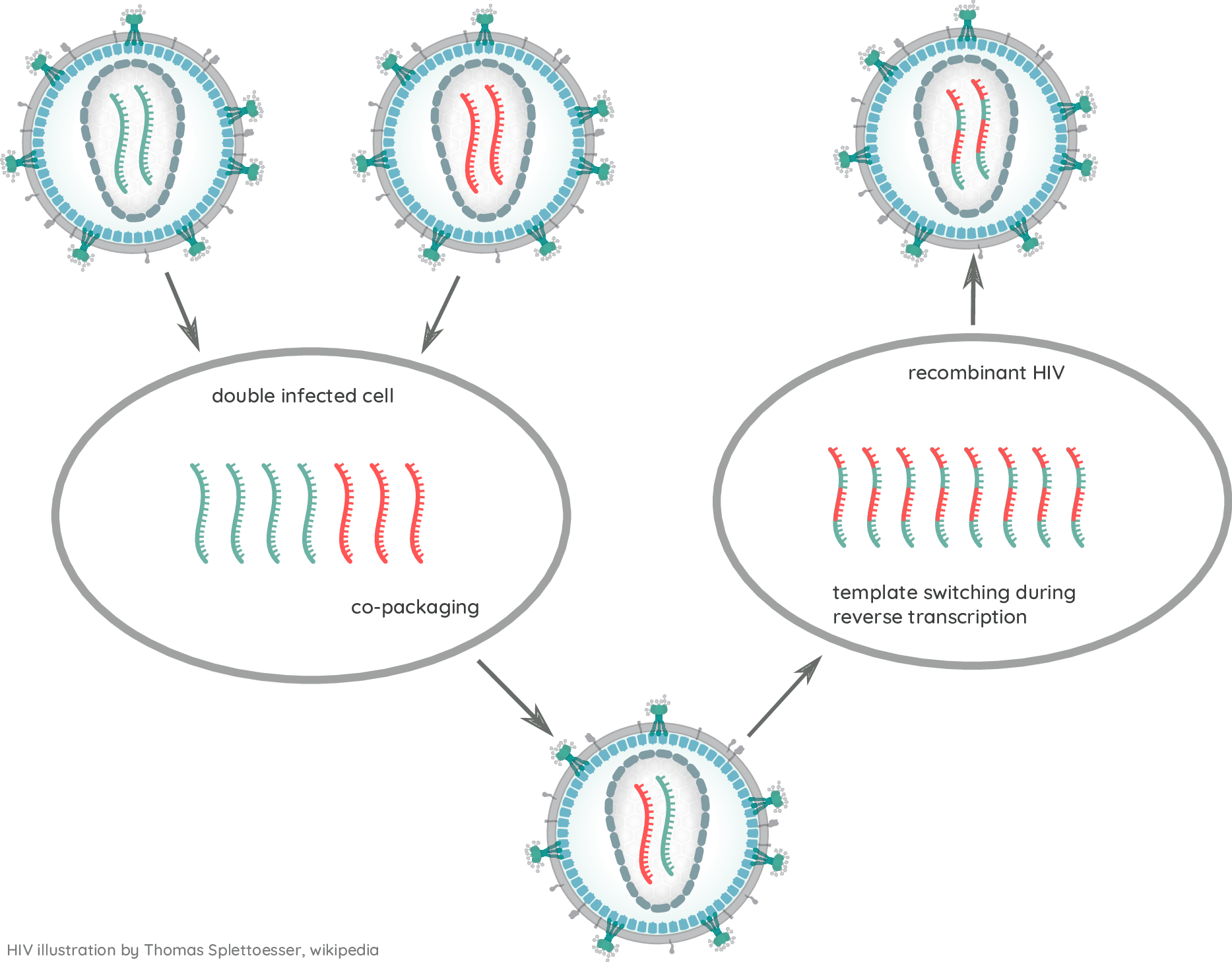
HIV-1 evolution within one individual
- Viruses rapidly diversify
→ swarm or quasi-species - Different tissues can harbor different viral variants
- diversity depends on duration of infection
- number of founder variants
- immune selection
Drug resistance evolution
Drug targets
- Reverse transcriptase (NRTI and NNRTI)
- Protease inhibitors
- Integrase inhibitors
- Entry inhibitors
Resistance evolution
- Every mutations pre-exists → mono-therapy fails
- Combination therapy requires multiple mutations to become resistant
- Sufficiently high 'barrier to resistance' results in long-term suppressive therapy
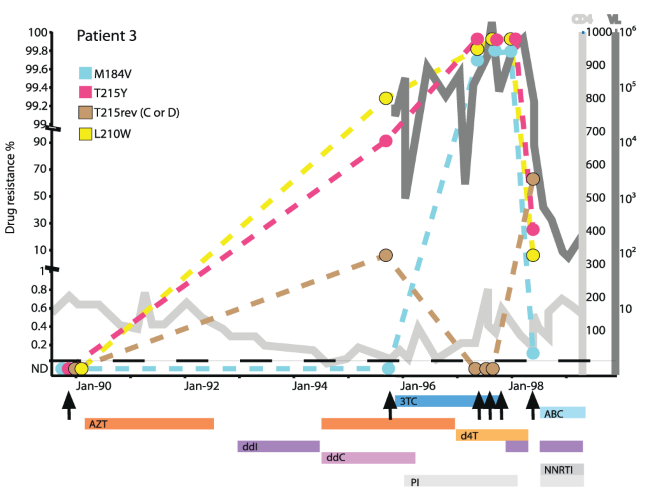
Sequence evolution vs phenotypic evolution
- RNA viruses change rapidly under consistent selection pressure
- Rapid immune escape or drug resistance in HIV or influenza
- BUT: most mutations are deleterious
- AND: most observed changes have no clear phenotype
Case in point: "the microcephaly mutation" in Zika
- Weak evidence that a particular mutation facilitates spread of Zika virus in mouse brains
- Media trumpets: "microcephaly causing mutation" found
- Zika virus strains in India lack this mutations
→ authorities claim: Zika not a problem - see Open-Ed by Nathan Grubaugh for a good discussion


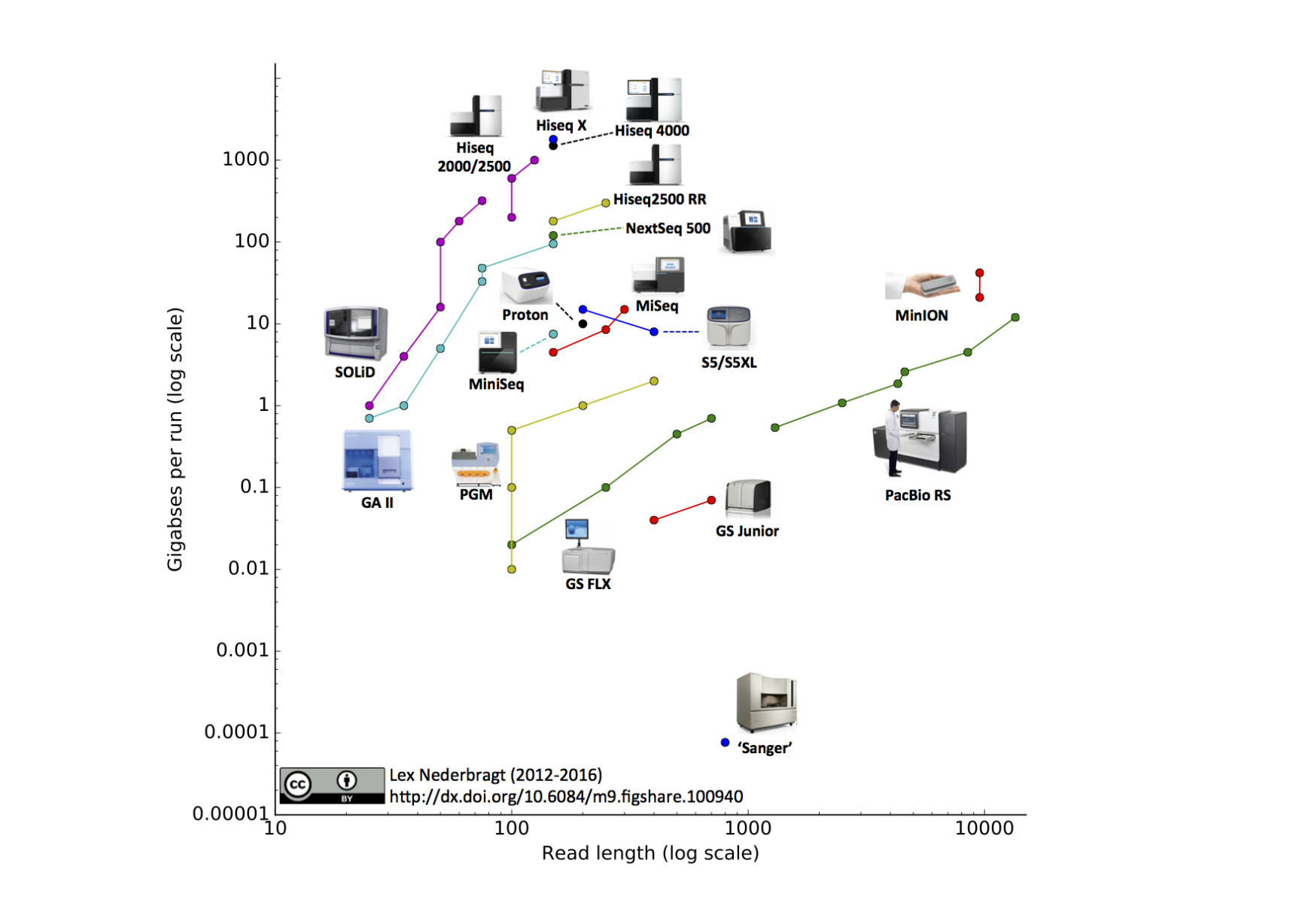
Development of sequencing technologies
We can now sequence...
- thousands of bacterial isolates
- thousands of single cells
- populations of pathogens, metagenomics