Establishment and maintenance of the latent HIV-1 DNA reservoir
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/201901_StLucia.html
The people who made this happen...
- Fabio Zanini
- Jan Albert
- Johanna Brodin
- Christa Lanz
- Göran Bratt
- Lina Thebo
- Vadim Puller



HIV-1 evolution within one individual
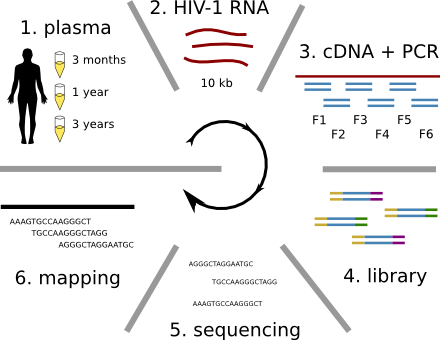

HIV-1 sequencing before and after therapy
 Zanini et al, eLife, 2015;
Brodin et al, eLife, 2016.
Collaboration with the group of Jan Albert
Zanini et al, eLife, 2015;
Brodin et al, eLife, 2016.
Collaboration with the group of Jan Albert
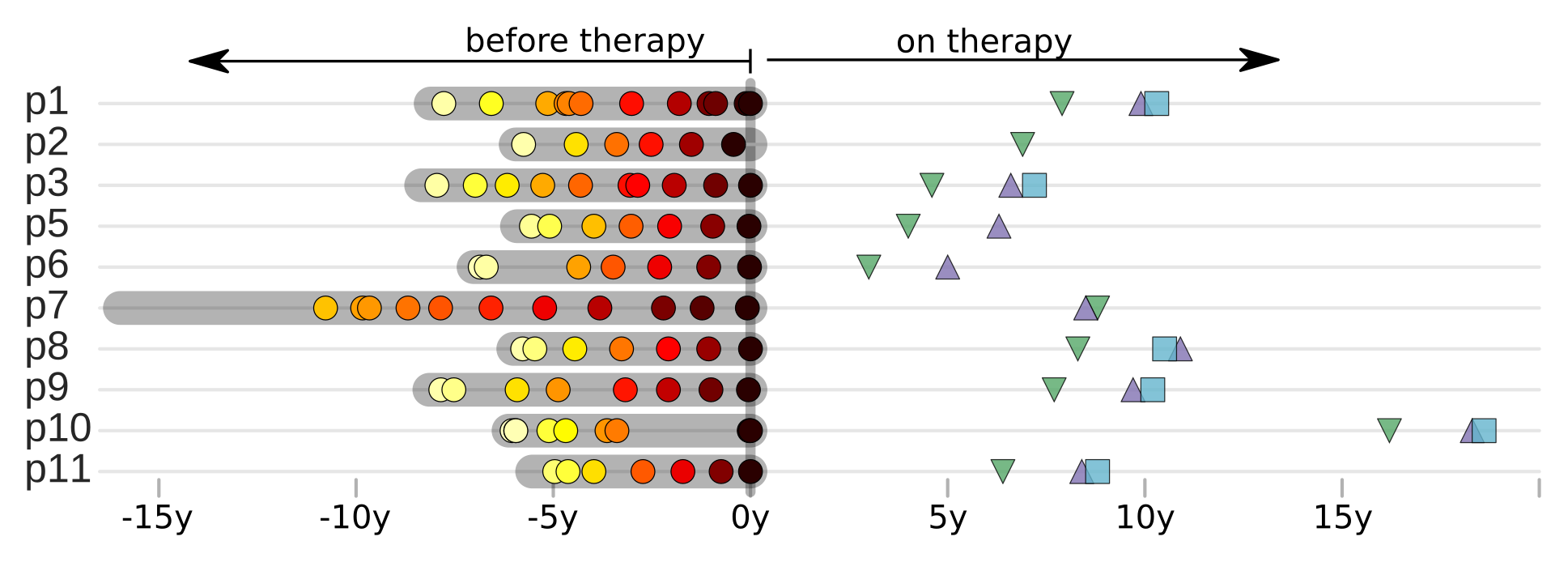
Population sequencing to track all mutations above 1%
Detailed time-resolved record of evolution of the entire genome (here V3 in patient 3)
Frequent reversion towards the center of HIV phylogeny
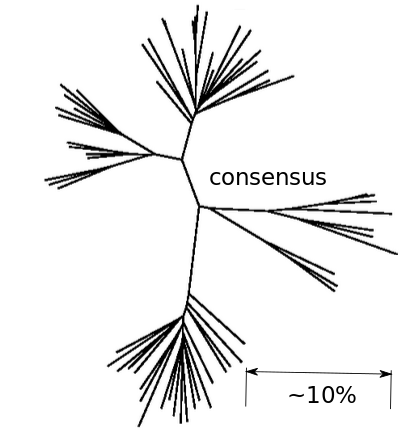
- Almost one third of all mutations are reversions (~5% expected by chance)
- Reversions accumulate slowly throughout chronic infection
- Reversion can explain much of the intra/inter-host evolutionary rate mismatch
Divergence at increasingly conserved positions
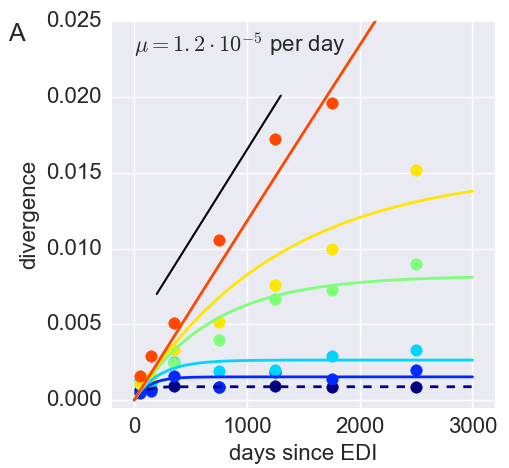
- Six categories from high to low conservation
- mutation away from preferred state with rate $\mu$
- selection against non-preferred state with strength $s$
- variant frequency dynamics: $\frac{d x}{dt} = \mu -s x $
- equilibrium frequency: $\bar{x} = \mu/s $
- fitness cost: $s = \mu/\bar{x}$
- Fit model of minor variation to categories of conservation
- $\Rightarrow$ harmonic average fitness cost in category
Fitness landscape of HIV-1
Zanini et al, Virus Evolution, 2017p17 sequencing from PBMCs on fully suppressive therapy
 Brodin et al, eLife, 2016
Brodin et al, eLife, 2016
Hypermutated and intact proviral p17 sequences
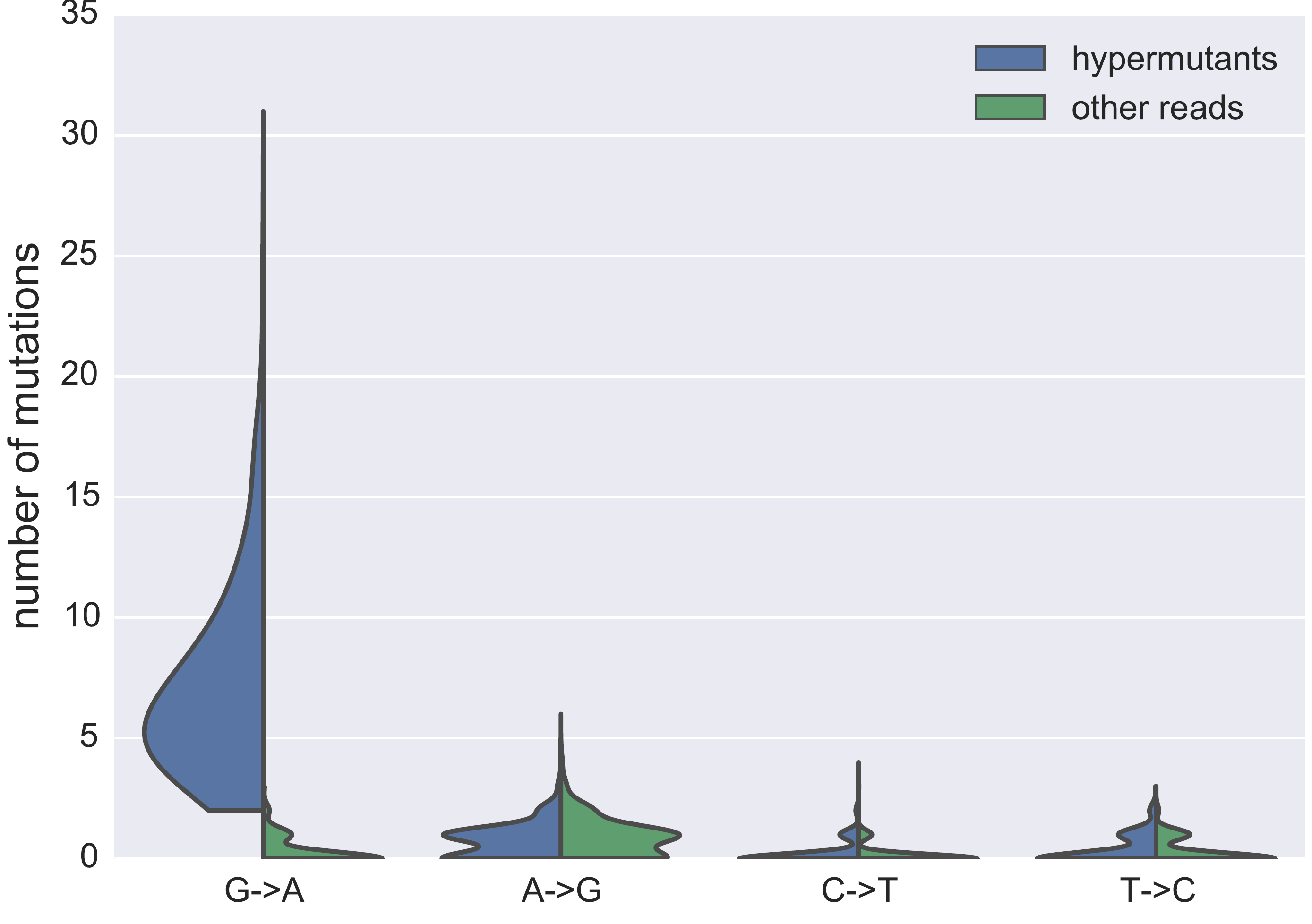
- Between 0 and 43% hypermutants, median 13%
- Between 3 and 43 unique sequences >1% read frequency, median 25
- Majority of hypermutant sequences have stop codons
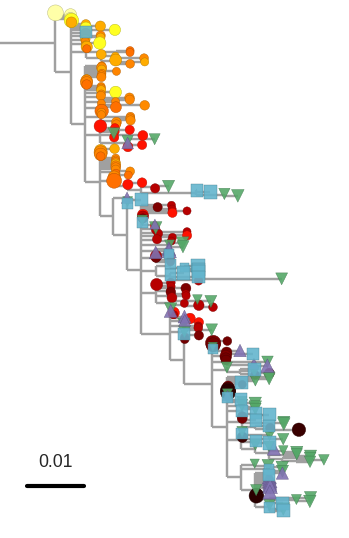
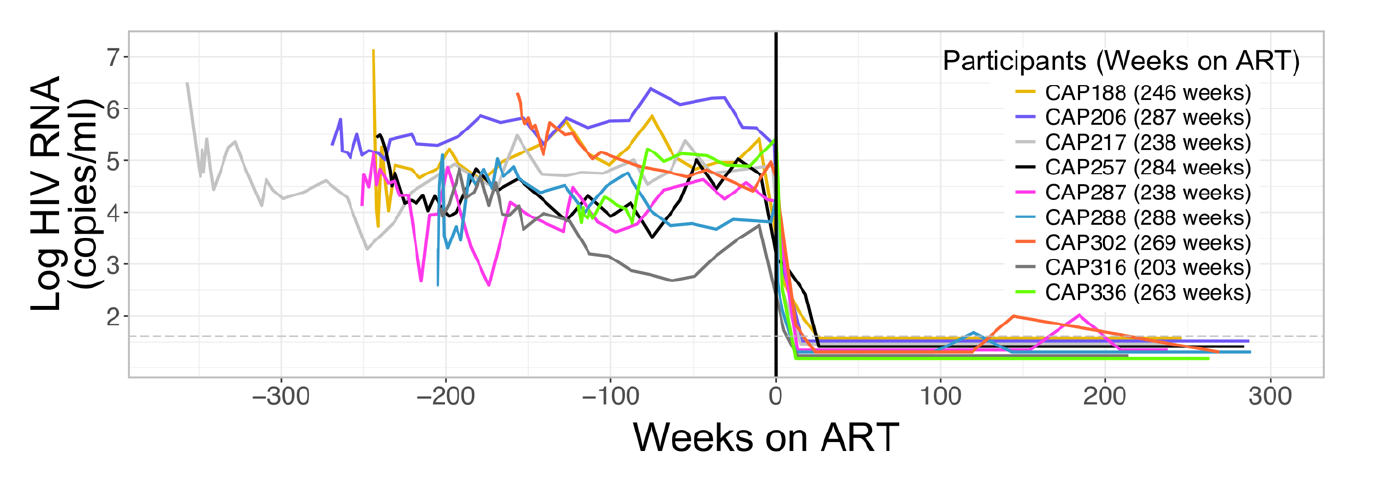
Proviral DNA mostly identical to pre-therapy RNA
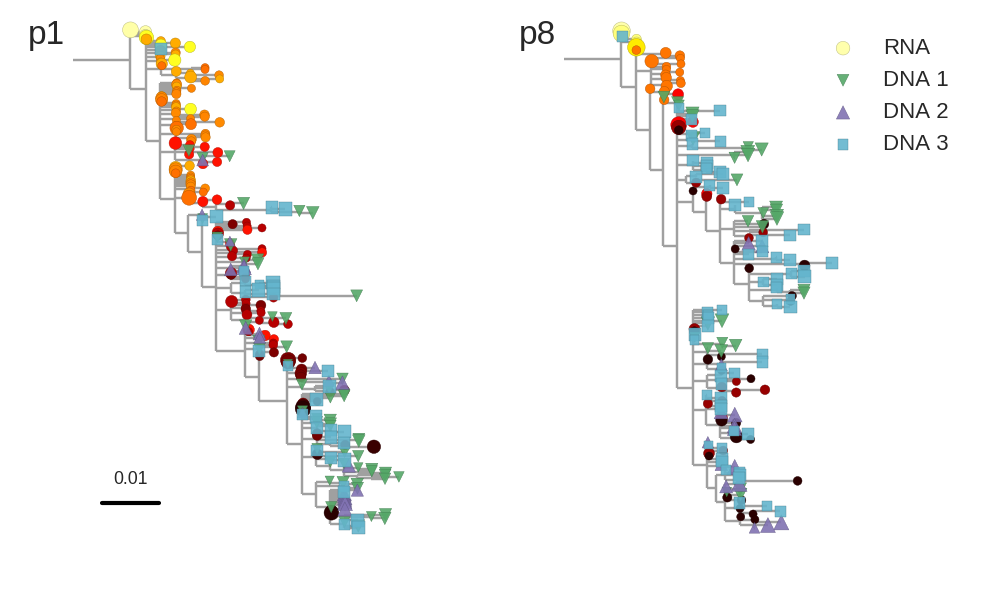
No evidence of ongoing evolution
No evidence of ongoing evolution -- hypermutated provirus
 Brodin et al, eLife, 2016
Brodin et al, eLife, 2016
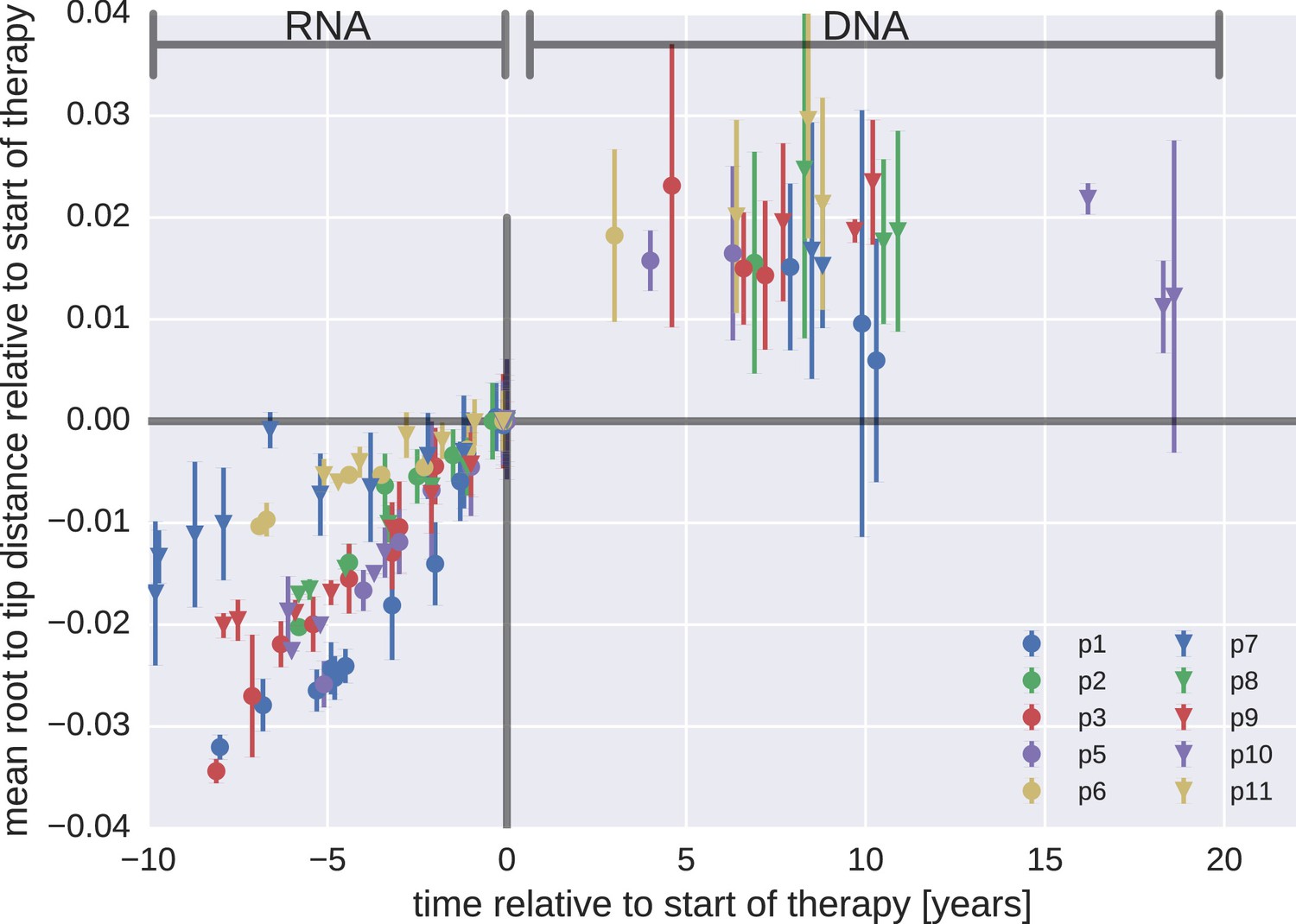
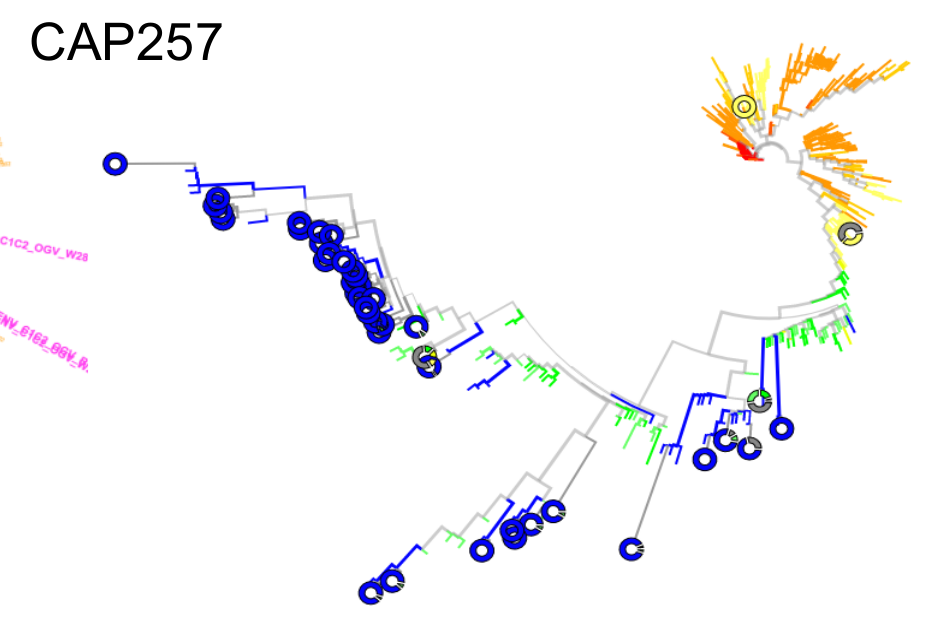
Most proviral DNA originates from shortly after start of therapy
- latent HIV → barcode of a T-cell lineage
- all latent integrated virus derives from late infection
Possible explanations for reservoir establishment time distributions
Reservoir shaped by T-cell turn-over
- T-cell lineages are short lived w/o treatment
- The reservoir is cleared/superseeded by subsequent infection
- on therapy: T-cell clones live decades
- pre-therapy state is frozen
Reservoir formation triggered by immune deactivation
- → Melissa Abrahams talk on Tuesday
- Abrahams et al, bioRxiv, 2019
- Similar study design, but functional outgrowth virus!
Summary
- Continuous reversion to ancestral states throughout chronic infection
- Minor diversity allows fitness cost estimates at every position in the genome
- The latent reservoir is a snapshot of the pre-treatment population
- No evidence of ongoing replication
- No clear trend of diversity loss on treatment
- Proviral DNA can serve as a T-cell lineage marker
Acknowledgments
- Fabio Zanini
- Jan Albert
- Johanna Brodin
- Christa Lanz
- Göran Bratt
- Lina Thebo
- Vadim Puller


