Tracking infectious disease and drug resistance using whole genome sequencing
Richard Neher
Biozentrum, University of Basel
Sequences record the spread of pathogens




Human seasonal influenza viruses




- Influenza viruses evolve to avoid human immunity
- Vaccines need frequent updates

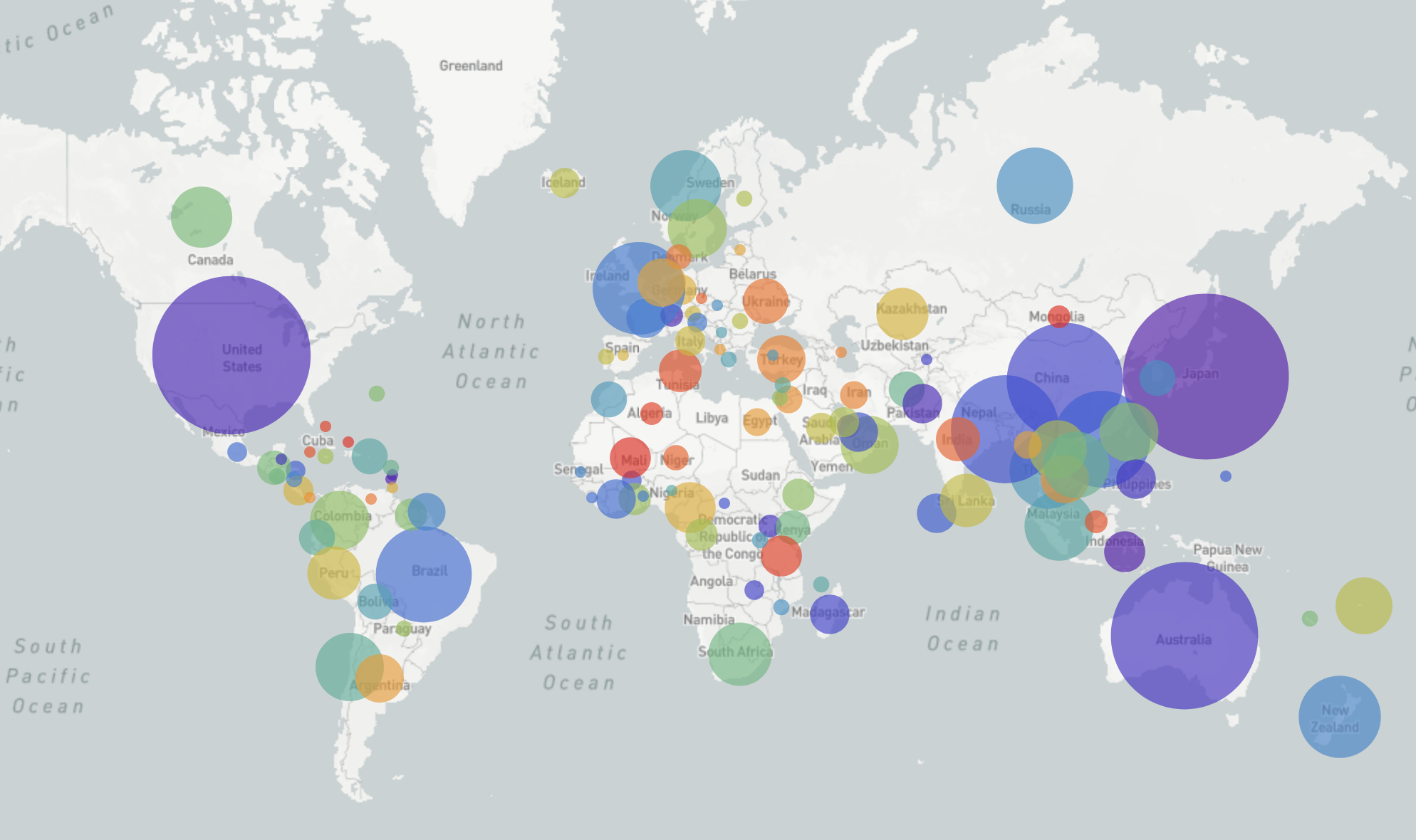
nextstrain.org
joint project with Trevor Bedford & his lab

Enterovirus D68 -- with Robert Dyrdak, Emma Hodcroft & Jan Albert
- Non-polio enterovirus
- Almost everybody has antibodies against EV-D68
- Large outbreak in 2014 with severe neurological symptoms in
young children (acute flaccid myelitis) - Another outbreak in 2016
- Outbreaks tend to start in late summer/fall
- Several reports of EV-D68 outbreaks last fall
(201 AFM cases in the US in 2018)
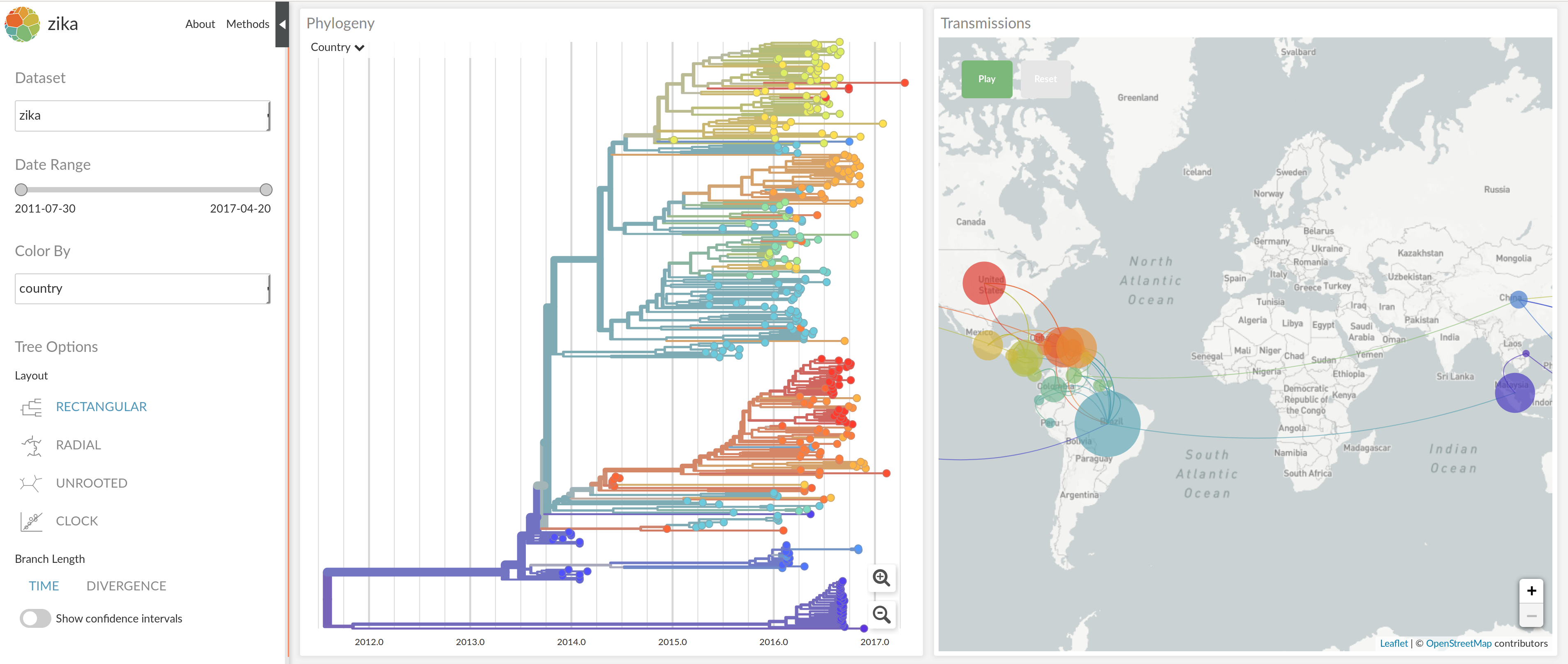
How does EV-D68 spread? Does it change?
nextstrain.org/enterovirus
joint work with Robert Dyrdak, Emma Hodcroft & Jan Albert
Whole genome deep sequencing of Enterovirus D68
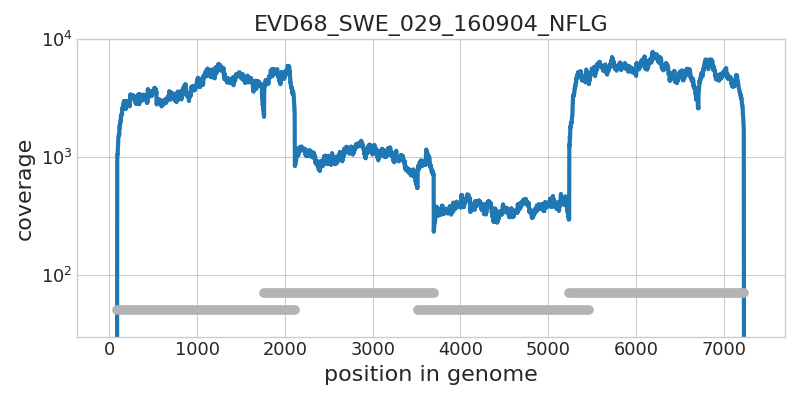
- Amplified in 4 overlapping segments
- Illumina sequenced to high coverage
iSNV frequency accuracy and sequencing errors
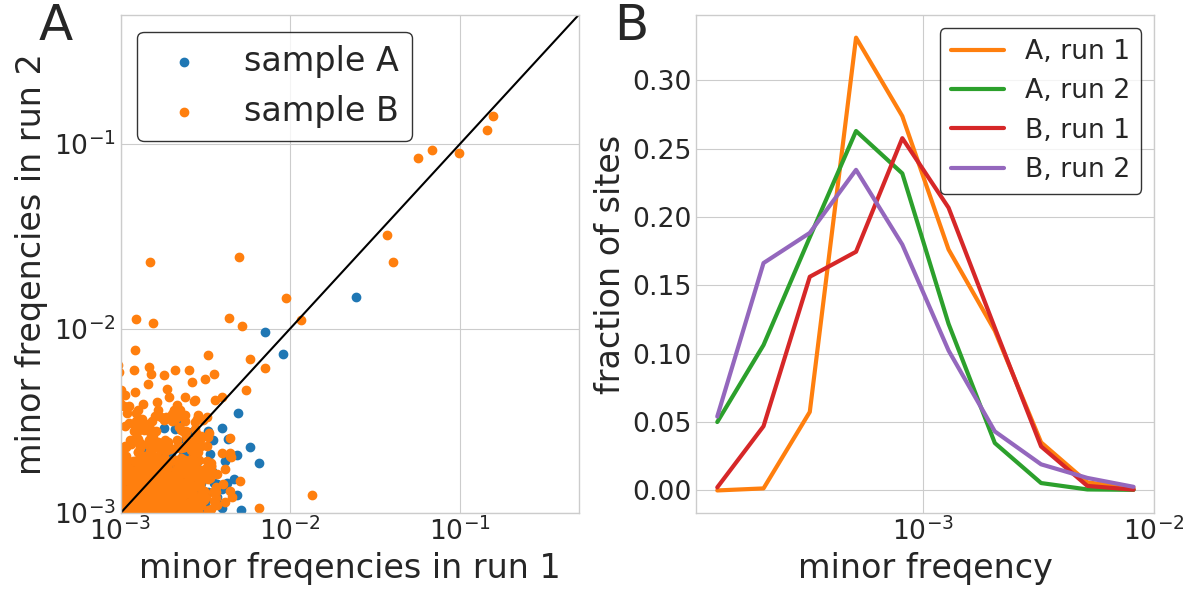
- iSNV frequencies reproducible above 1%
- background at around 1/1000
Infections with multiple variants
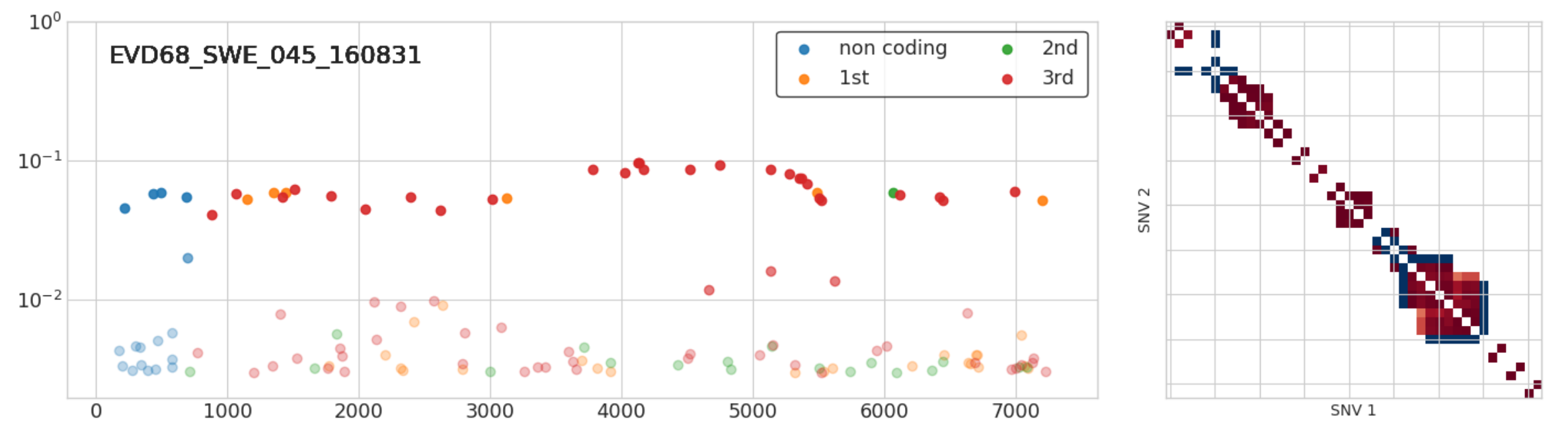
- A set of iSNVs at very similar frequencies in full linkage
- Suggest infection with two related variants
- 3 out of 50 samples: Implies high prevalence
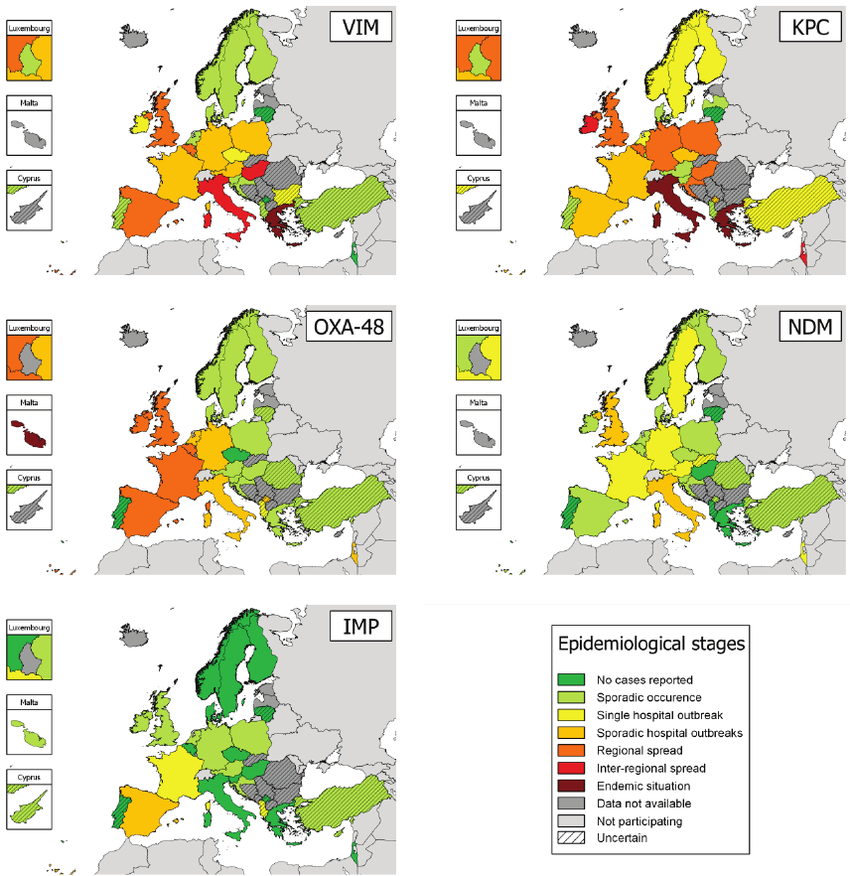
Carbapenemase producing bacteria
- Reserve antibiotics used to treat MDR bacteria
- Introduced in the 1980ies
- Resistance spread rapidly
- Resistance is mediated by several distinct beta-lactamases
→ pressing public health problem
→ fascinating instance of genes sweeping the globe by horizontal transfer
Tracking bacteria by sequencing
- Illumina → millions of short reads (<500bp)
- Too short to bridge repetitive elements
- → assemblies are fragmented into 100s of "contigs"
- Problem: all the important bits are flanked by repetitive/mobile elements

(really terrible example)
Long-read sequencing



Long-read sequencing of Carbapenemase producing bacteria
- Contigs with drug resistance genes ~1-6 genes
→ no phylogenetic resolution - long-read assemblies give full length plasmids
- tracking via synteny and structural diversity, not SNPs
→ we need to reconstruct spread from genome structure evolution
Synteny alignments of Carbapenemase containing loci
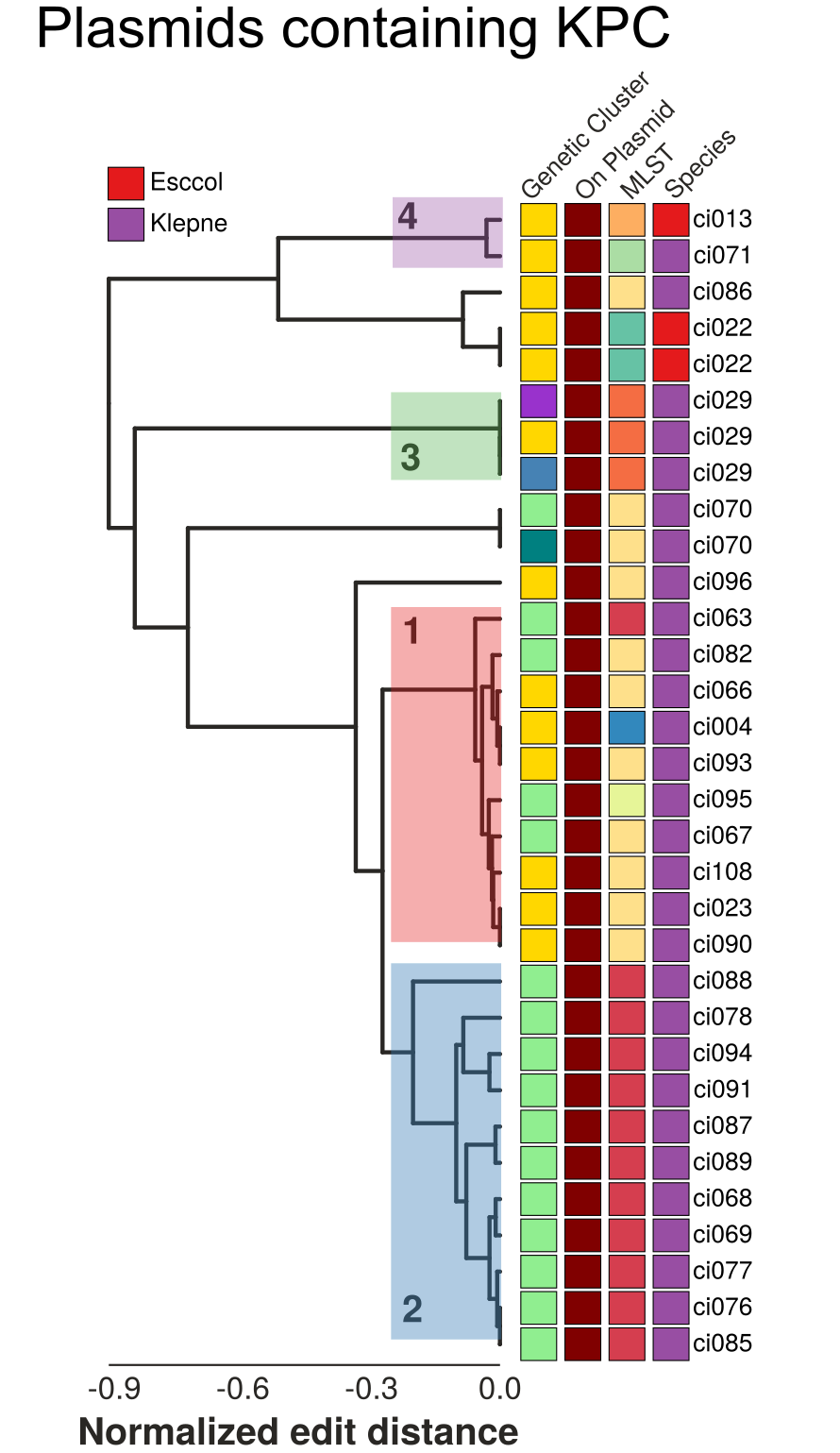
- Structural changes resolve evolutionary relationships
- Different KPC alleles are found on the same backgrouond
- Identical KPC alleles are found on different backgrounds
- Similar plasmids are spread across MLSTs and species boundaries
Synteny alignments of Carbapenemase containing loci

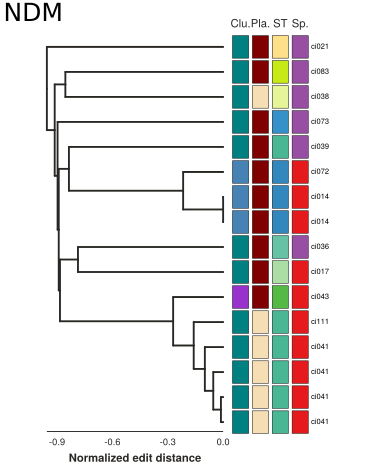
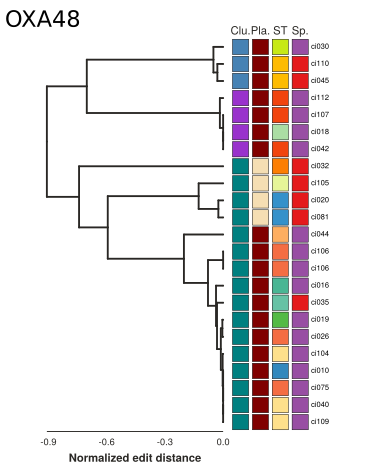
Summary
- Timely data sharing + automated analysis allows near real-time tracking of influenza
- Such analyses provide important input for vaccine strain selection
- Sequencing, analysis, and dissemination can be rapidly set-up for emerging pathogens
- Bacterial pathogens come with a special set of challenges
- Fascinating instance of evolution beyond SNPs
Acknowledgments -- nextstrain






- Trevor Bedford
- Colin Megill
- Pavel Sagulenko
- Sidney Bell
- James Hadfield
- Wei Ding
- Emma Hodcroft
- Sanda Dejanic
- John Huddleston
- Barney Potter




Acknowledgments -- Enterovirus
- Robert Dyrdak
- Jan Albert
- Lina Thebo
- Emma Hodcroft
- Bert Niesters (Groningen)
- Randy Poelman (Groningen)
- Elke Wollants (Leuven)

Acknowledgments -- Bacteria
- Wei Ding
- Nicholas Noll
- Eric Ulrich
- Adrian Egli (at USB)

With-in host diversity
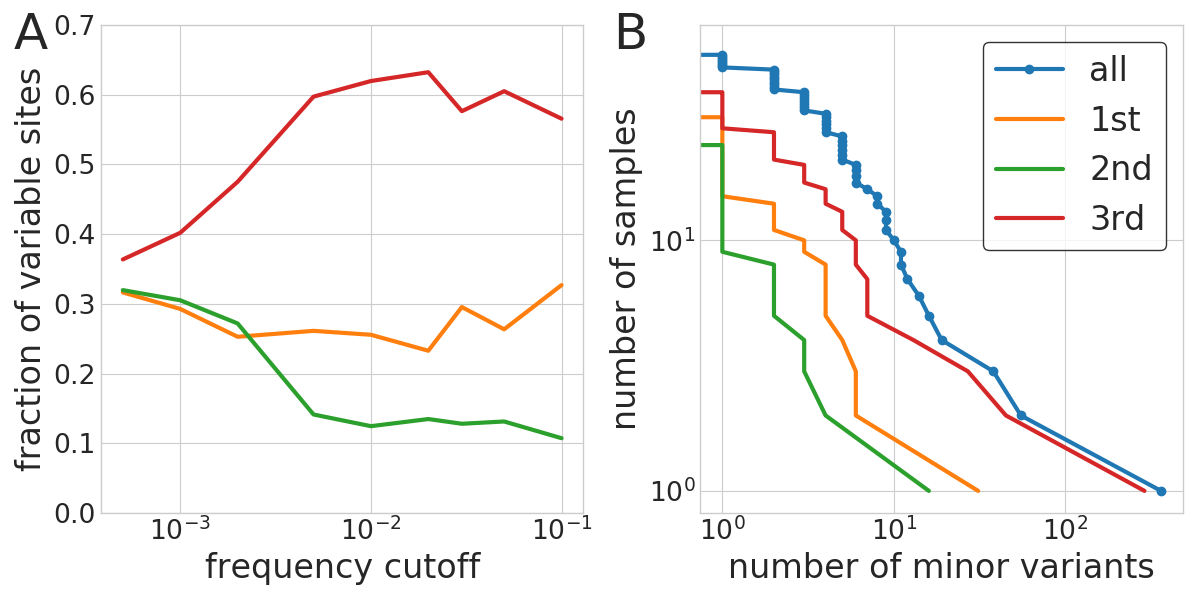
- Above 0.5%, iSNVs are biological
- Most samples have few iSNVs, three had more than 20