RNA virus evolution and the predictability of next year's flu
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/201903_APS.html
Human seasonal influenza viruses

Sequences record the spread of pathogens







- Influenza virus evolves to avoid human immunity
- Vaccines need frequent updates

Vaccine strain selection schedule
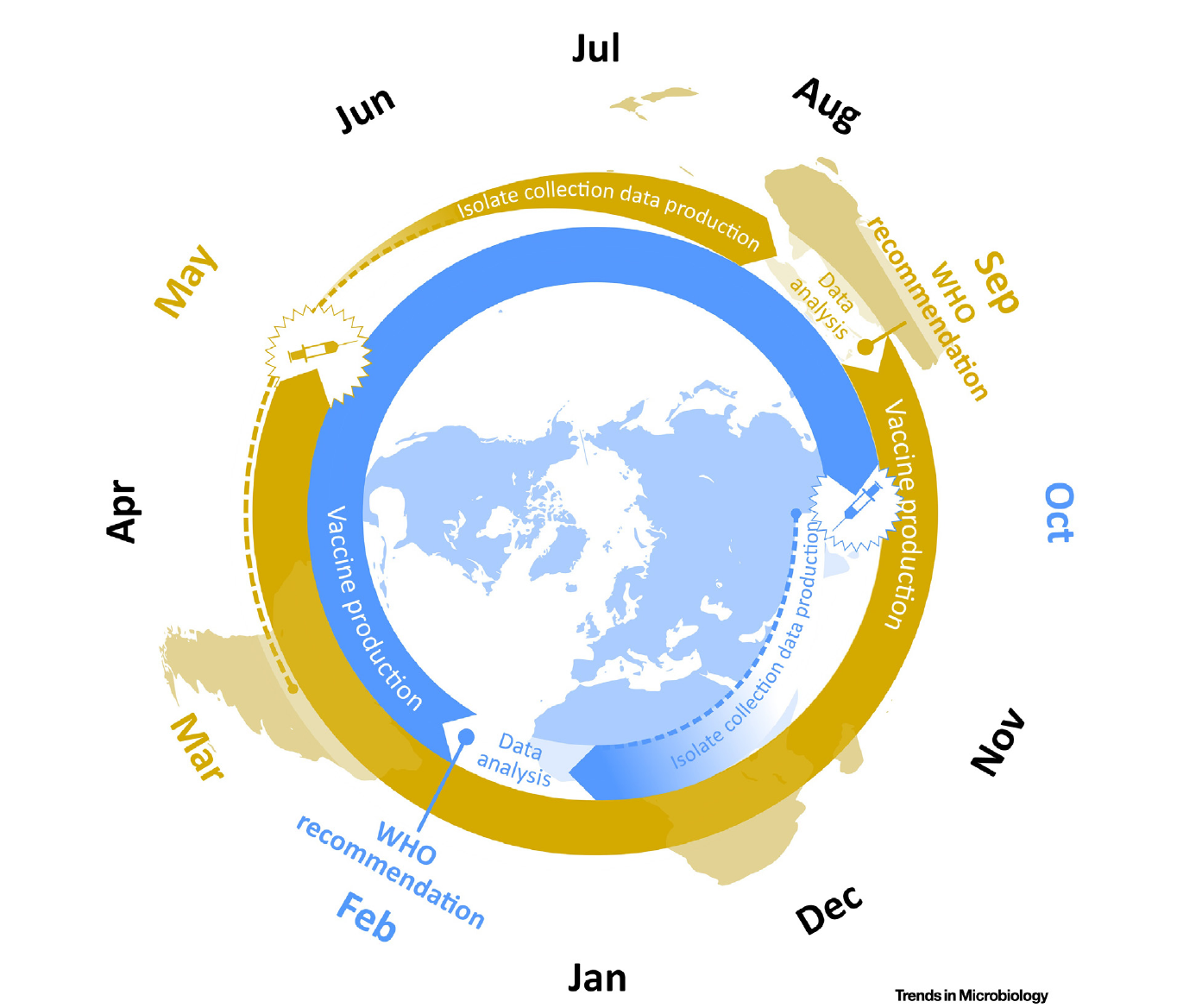 Klingen and McHardy, Trends in Microbiology
Klingen and McHardy, Trends in Microbiology
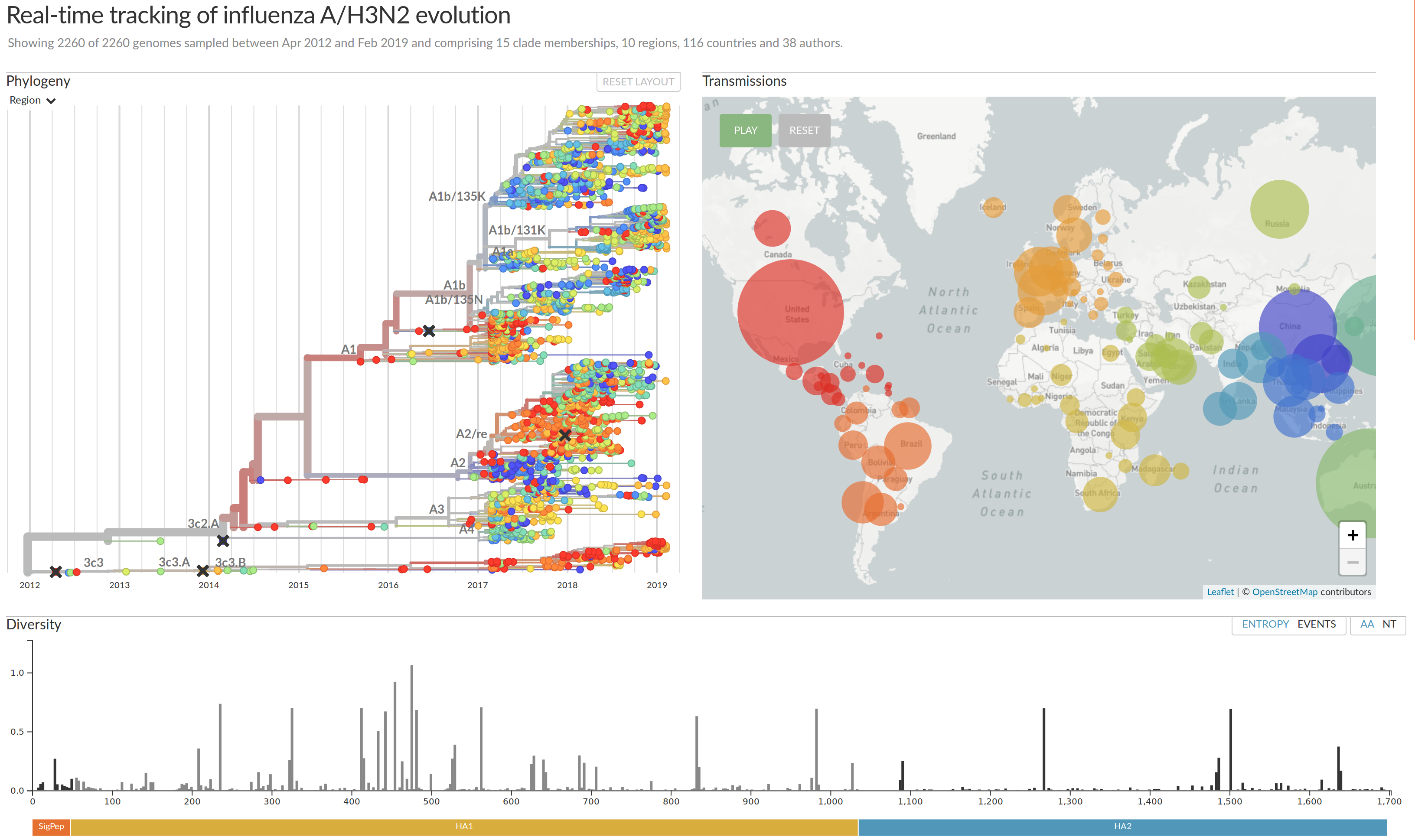
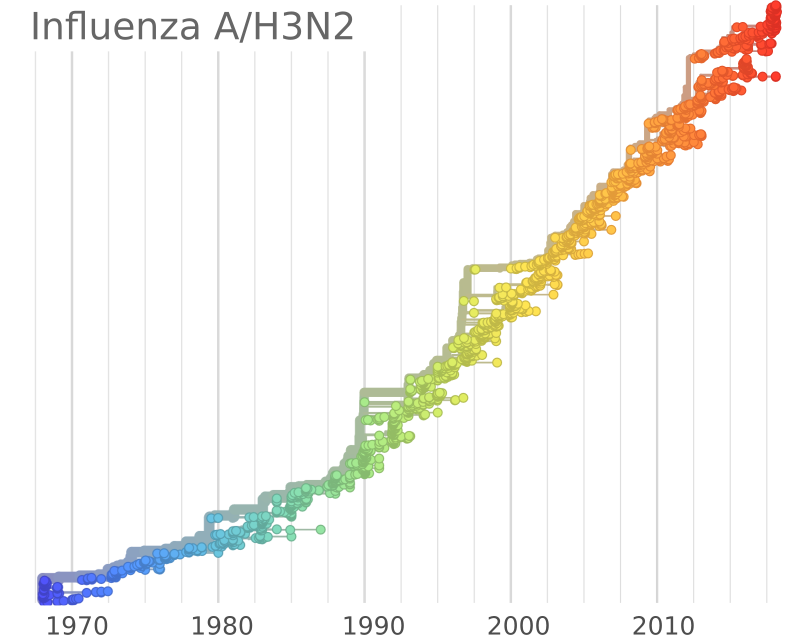
nextflu.org
joint work with Trevor Bedford & his lab
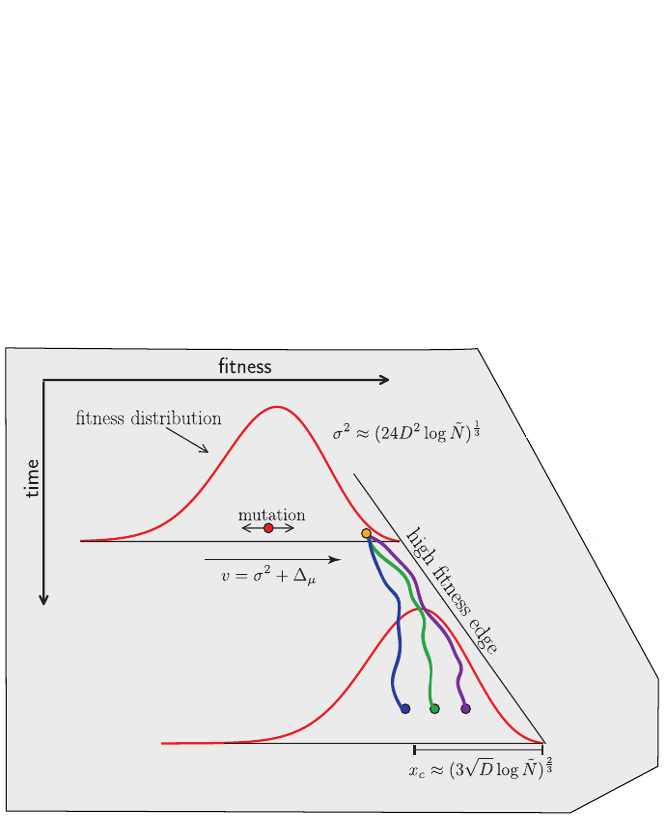
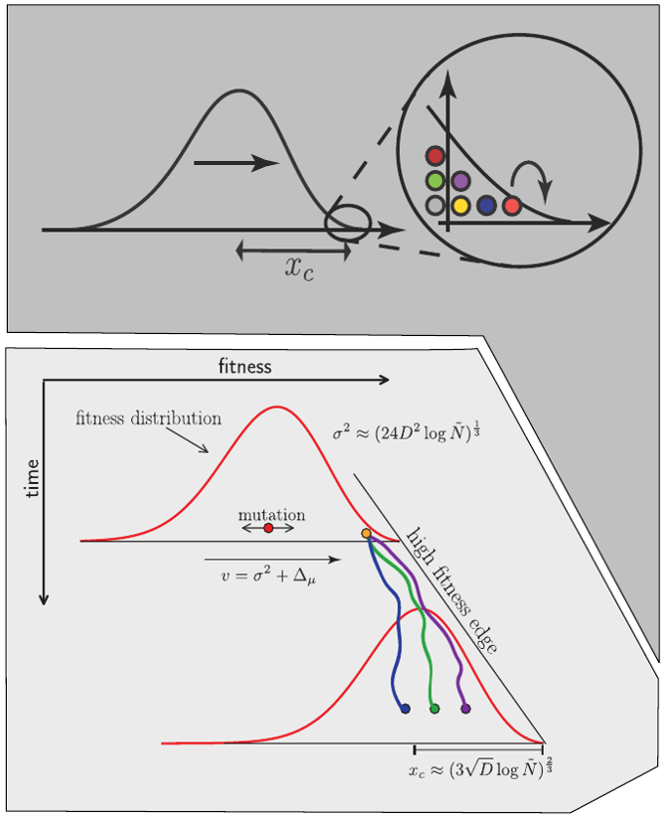
Fitness variation in rapidly adapting populations
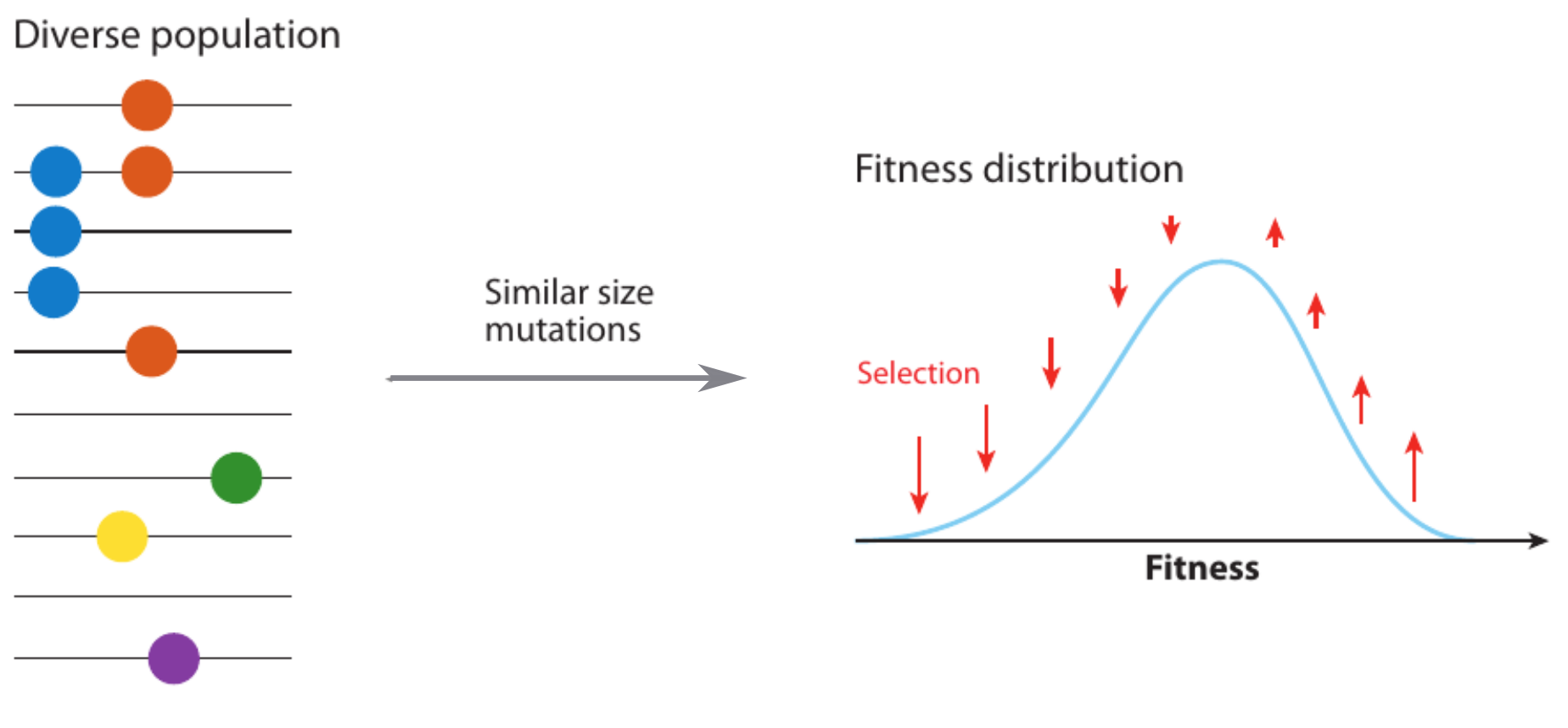
- Speed of adaptation is logarithmic in population size
- Environment (fitness landscape), not mutation supply, determines adaptation
- Different models have universal emerging properties
Neutral/Kingman coalescent
strong selection
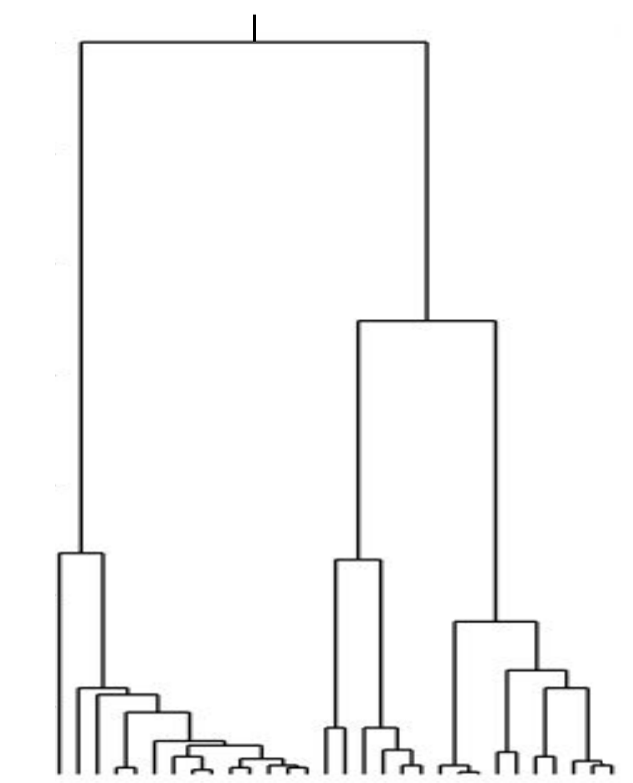

Bolthausen-Sznitman Coalescent
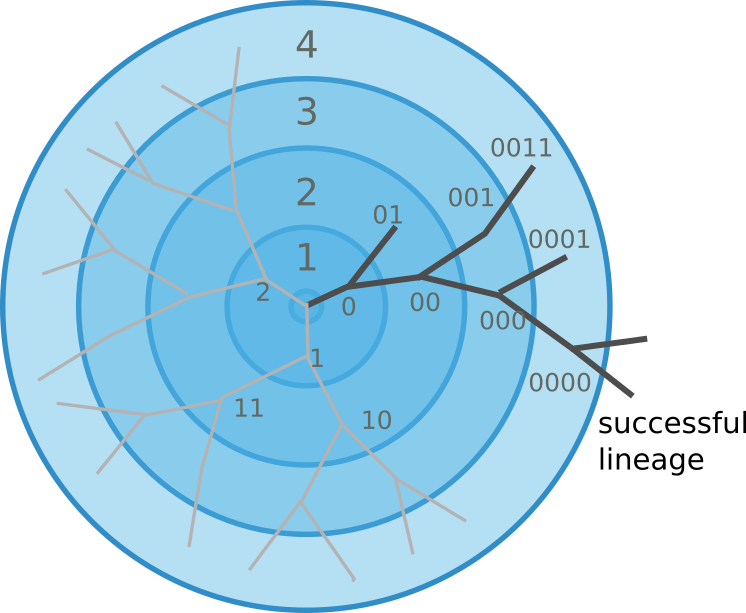
Bursts in a tree ↔ high fitness genotypes
Can we read fitness of a tree?
Predicting evolution

Given the branching pattern:
- can we predict fitness?
- pick the closest relative of the future?
Fitness inference from trees
$$P(\mathbf{x}|T) = \frac{1}{Z(T)} p_0(x_0) \prod_{i=0}^{n_{int}} g(x_{i_1}, t_{i_1}| x_i, t_i)g(x_{i_2}, t_{i_2}| x_i, t_i)$$
RN, Russell, Shraiman, eLife, 2014
Validate on simulation data
- simulate evolution
- sample sequences
- reconstruct trees
- infer fitness
- predict ancestor of future
- compare to truth

Validation on simulated data
Prediction of the dominating H3N2 influenza strain
- no influenza specific input
- how can the model be improved? (see model by Luksza & Laessig)
- what other context might this apply?
There are many ways to escape immunity -- why doesn't influenza speciate?

There are many ways to escape immunity -- why doesn't influenza speciate?
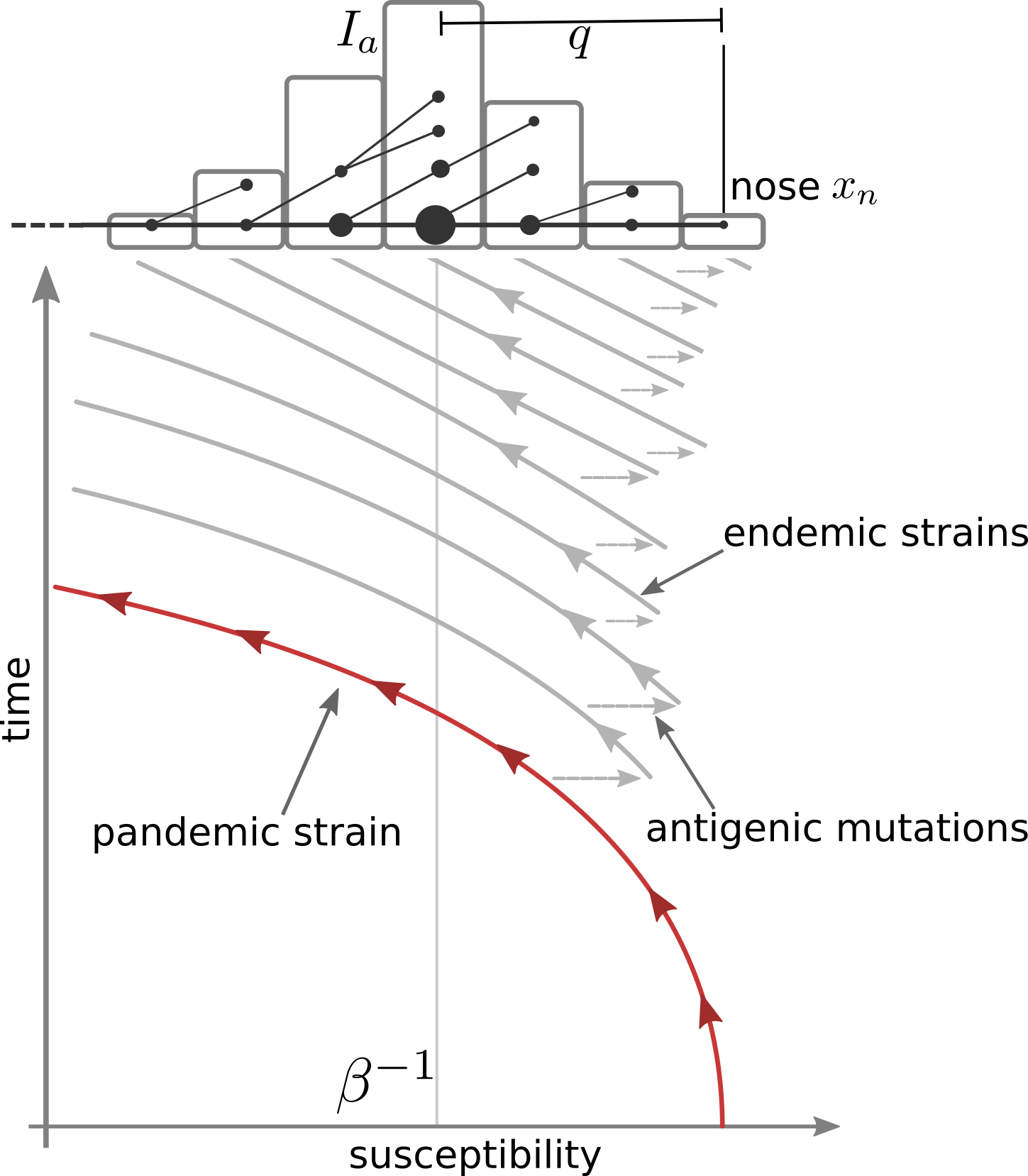
Combining SIR-models and rapid molecular adaptation
- Infections with strain $a$: $\frac{d I_a}{dt} = \beta S_a I_a - (\nu+\gamma)I_a$
- Susceptibility to strain $a$: $S_a =e^{-\sum_b K_{ab} R_b }$
- Recovered from stain $a$: $\frac{d R_a}{dt} = \nu I_a - \gamma R_a$
- Cross-immunity: $K_{ab} = e^{-\frac{|a-b|}{d}}$
- Mutations from $a\to b$ reduce cross-immunity and increase susceptibility
- Antigenic evolution is essential to establish seasonal patterns
Transition from pandemic to seasonal patterns
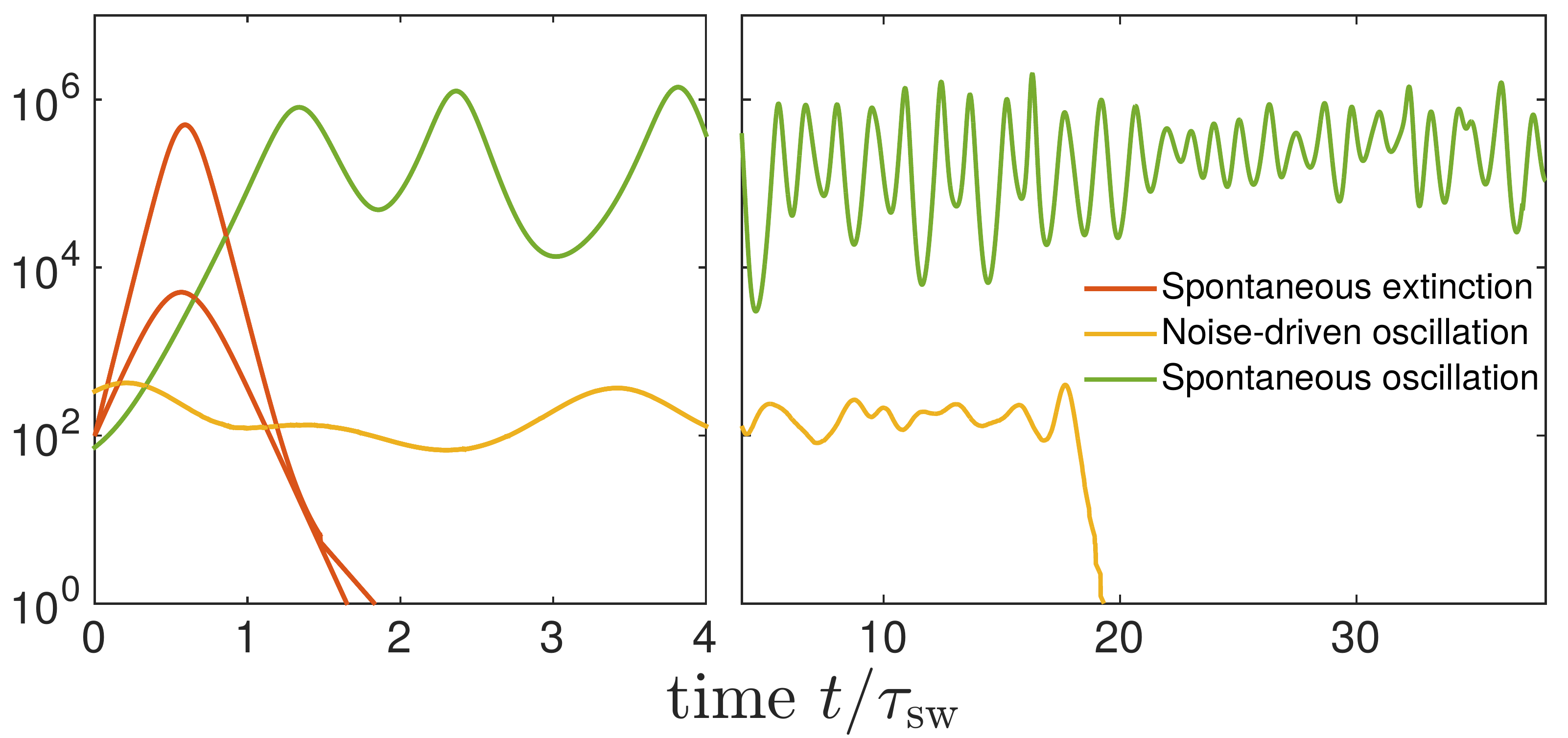
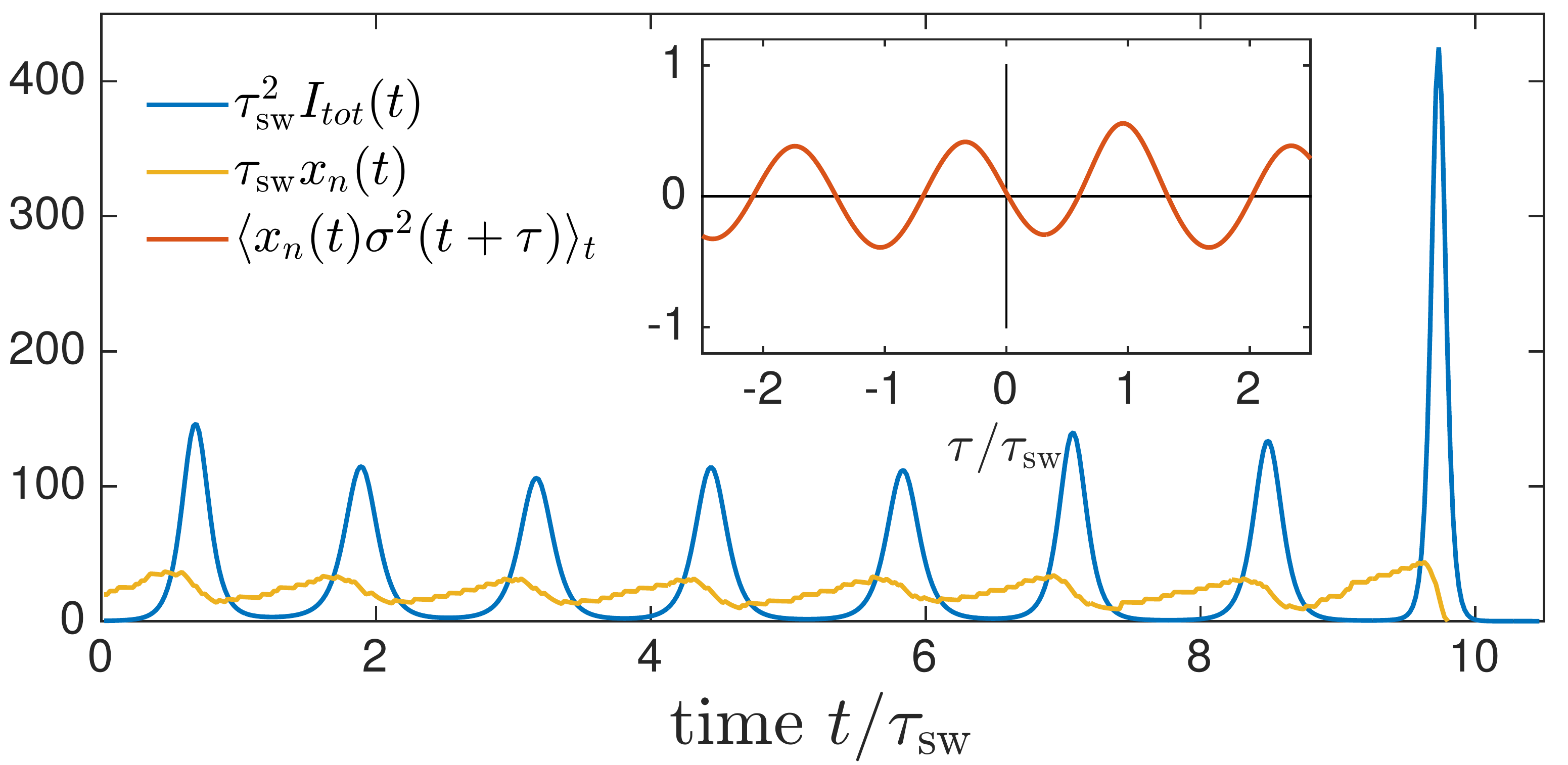
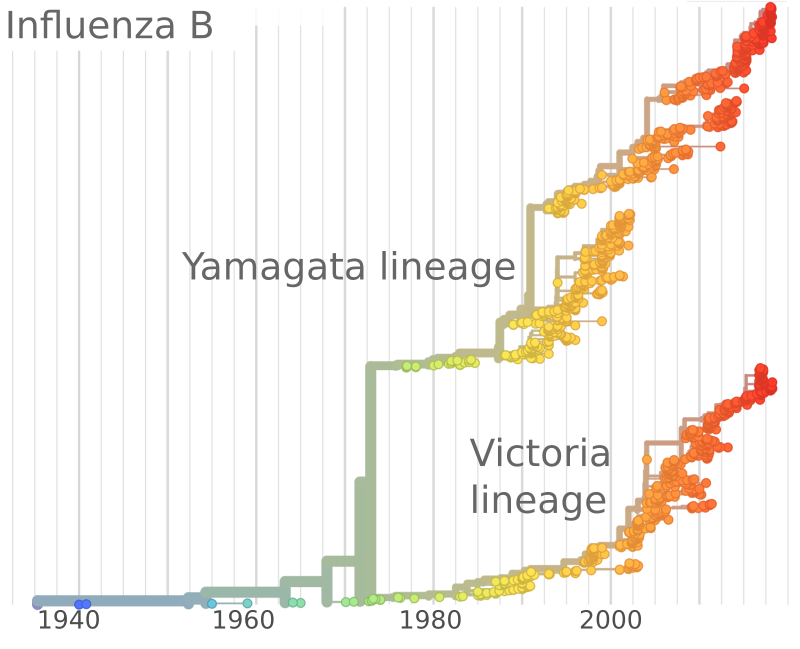
Speciation into antigenically distinct lineages
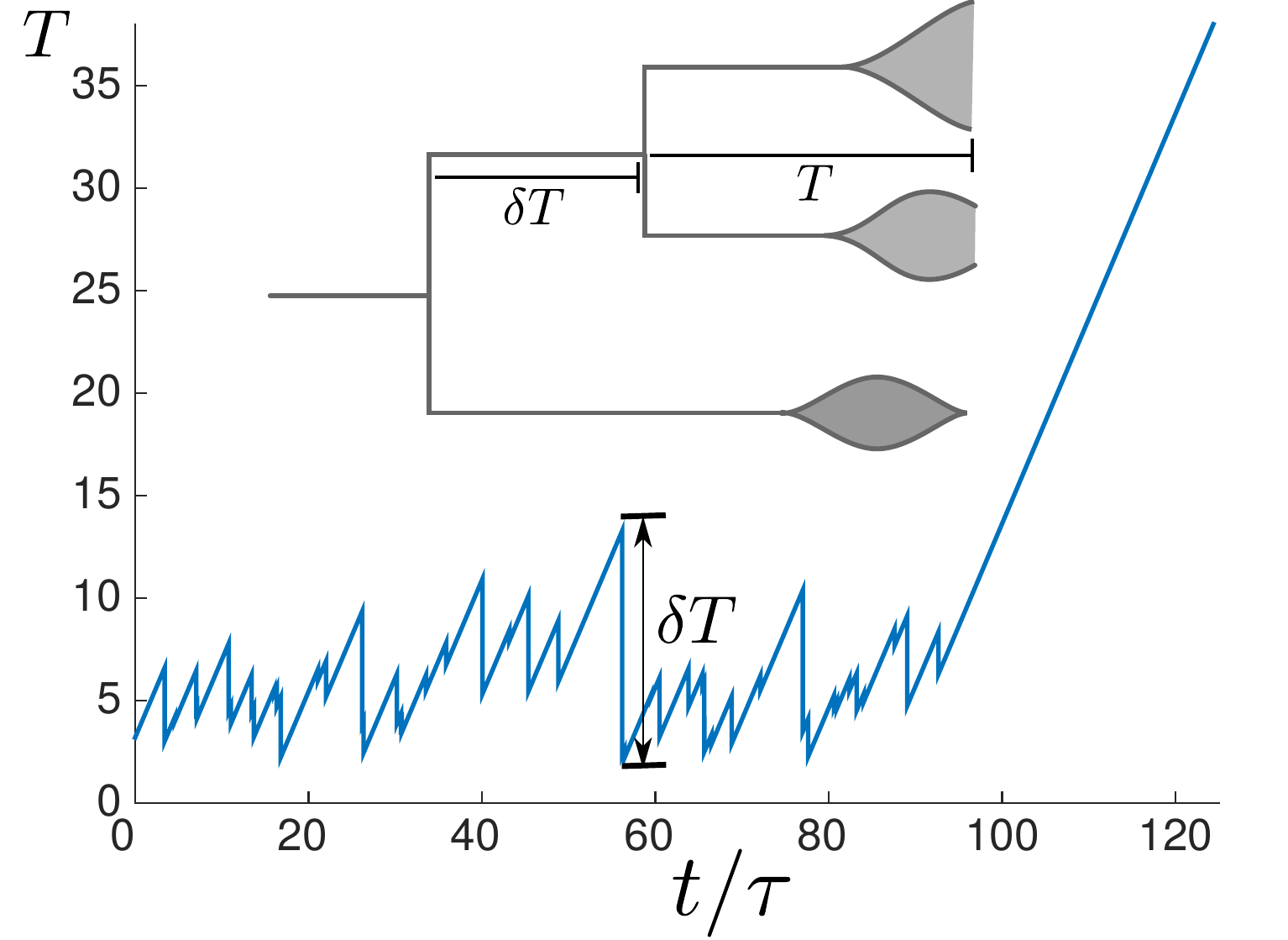
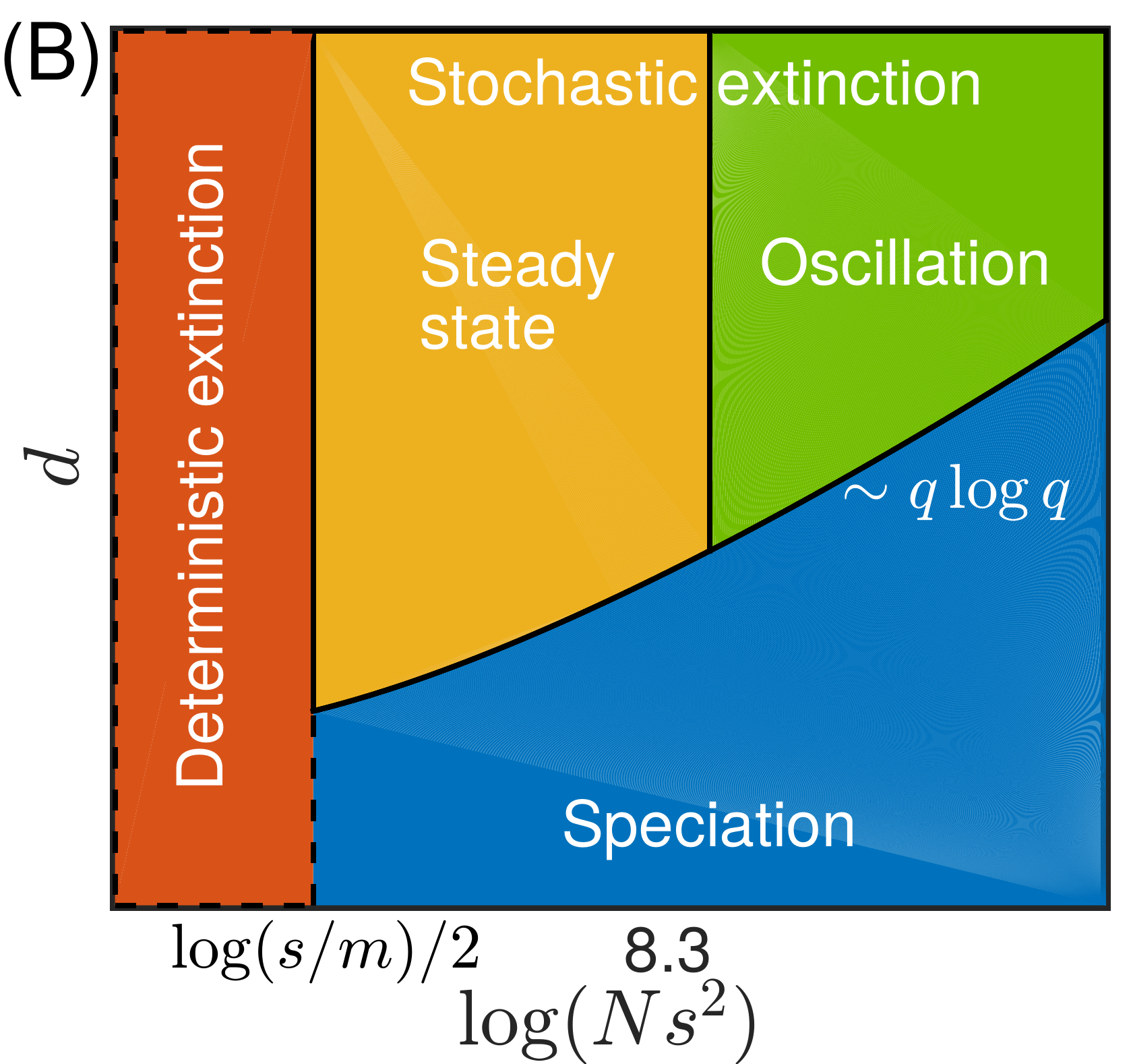
Summary
- RNA virus evolution can be observed directly
- Rapidly adapting population require new population genetic models
- Those model can be used to infer fit clades
- Future influenza population can be anticipated
- Automated real-time analysis can help fight the spread of disease
- Combining epidemiological and population genetic models can explain flu phylogenies
Influenza and Theory acknowledgments





- Le Yan
- Boris Shraiman
- Colin Russell
- Trevor Bedford
- Oskar Hallatschek



Acknowledgments -- nextstrain






- Trevor Bedford
- Colin Megill
- Pavel Sagulenko
- Sidney Bell
- James Hadfield
- Wei Ding
- Emma Hodcroft
- Sanda Dejanic
- John Huddleston
- Barney Potter



