Real-time tracking of pathogen evolution and spread
Richard Neher
Biozentrum & SIB, University of Basel
slides at neherlab.org/201907_ECDC.html
Sequences record the spread of pathogens




Mutations accumulate at a rate of $10^{-5}$ per site and day!
Influenza virus genome - 8 segments
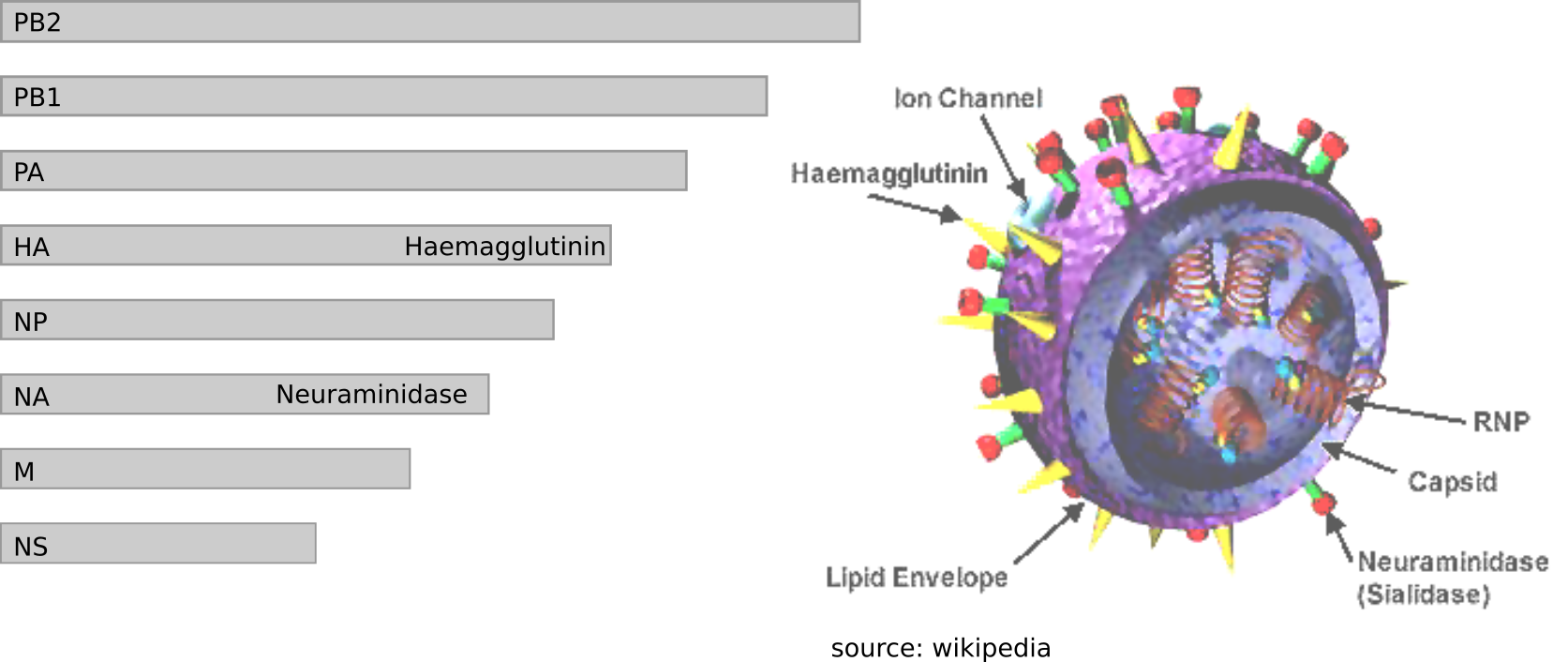
Zika virus genome $\sim 10000$ bases

Ebola virus genome $\sim 20000$ bases

Many RNA viruses pick up one mutation every 2-4 weeks!
Frequent mutations imply...
- most viruses in an outbreak differ from each other
- transmission chains are can be inferred
- transmission can be ruled out!
- geographic spread can be reconstructed
- phenotypic changes can be inferred (for some pathogens)
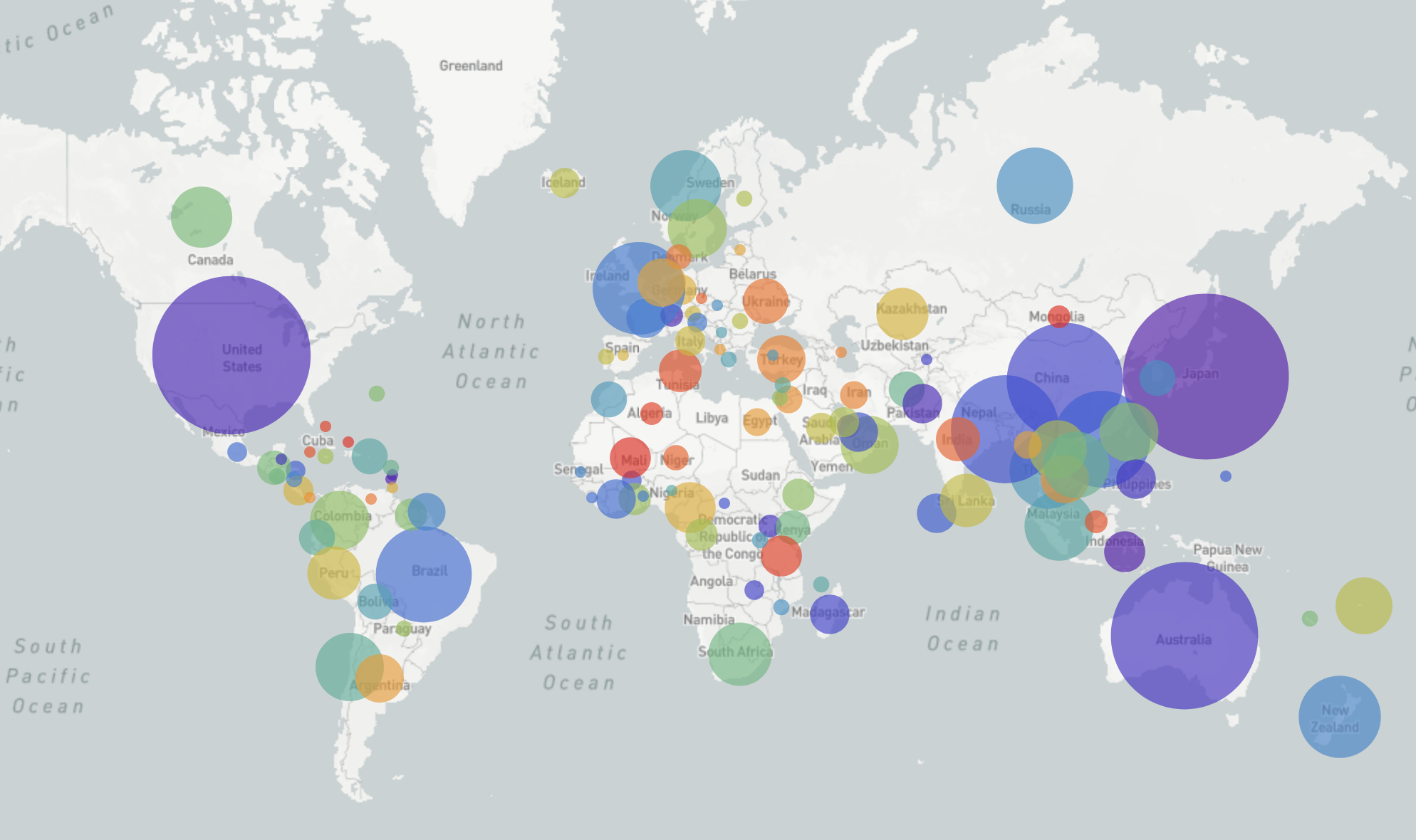
GISRS and GISAID -- Influenza virus surveillance
- comprehensive coverage of the world
- timely sharing of data -- often within 2-3 weeks of sampling
- hundreds of sequences per week (in peak months)
→ requires continuous analysis and easy dissemination
→ interpretable and intuitive visualization
nextflu.org
joint project with Trevor Bedford & his lab
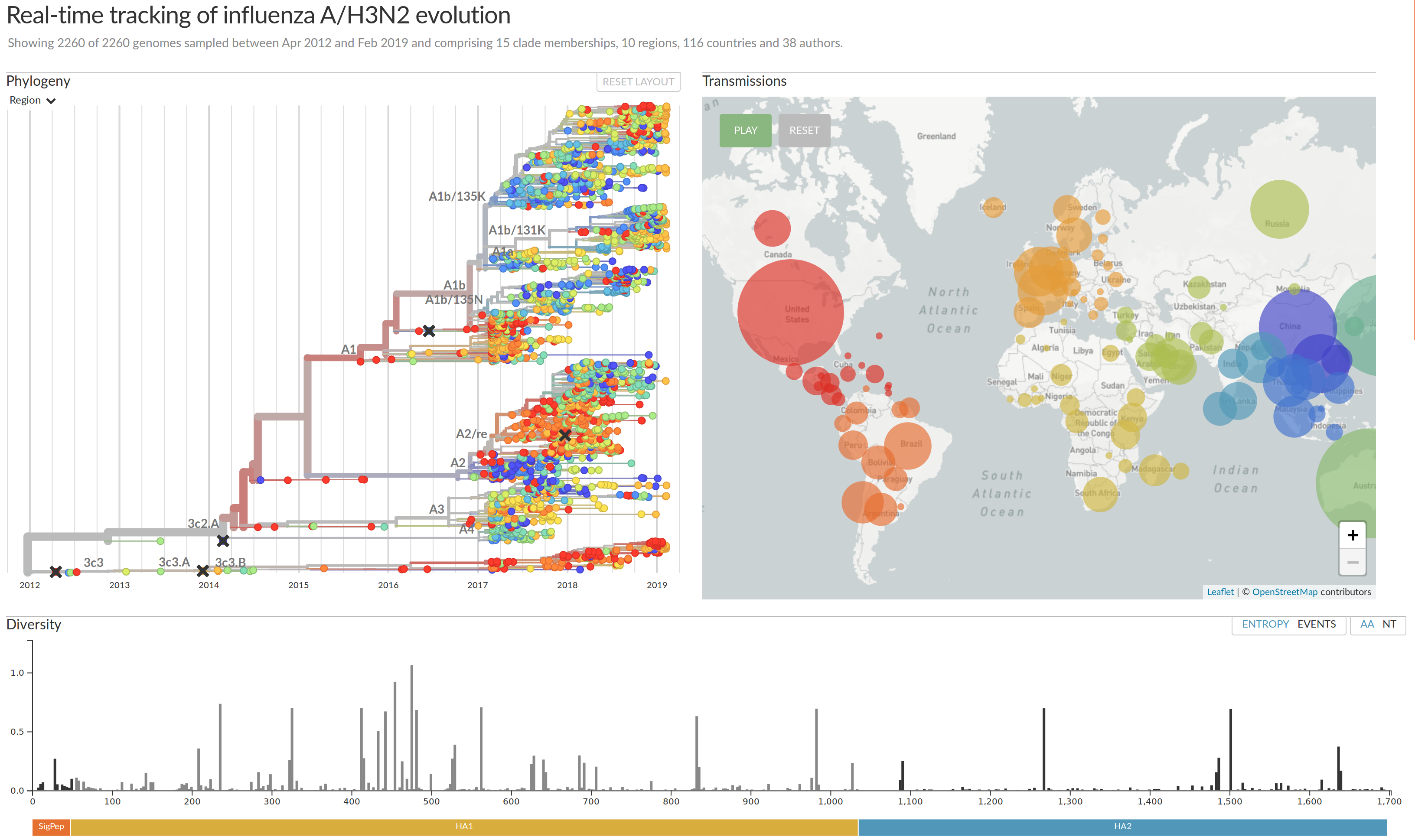
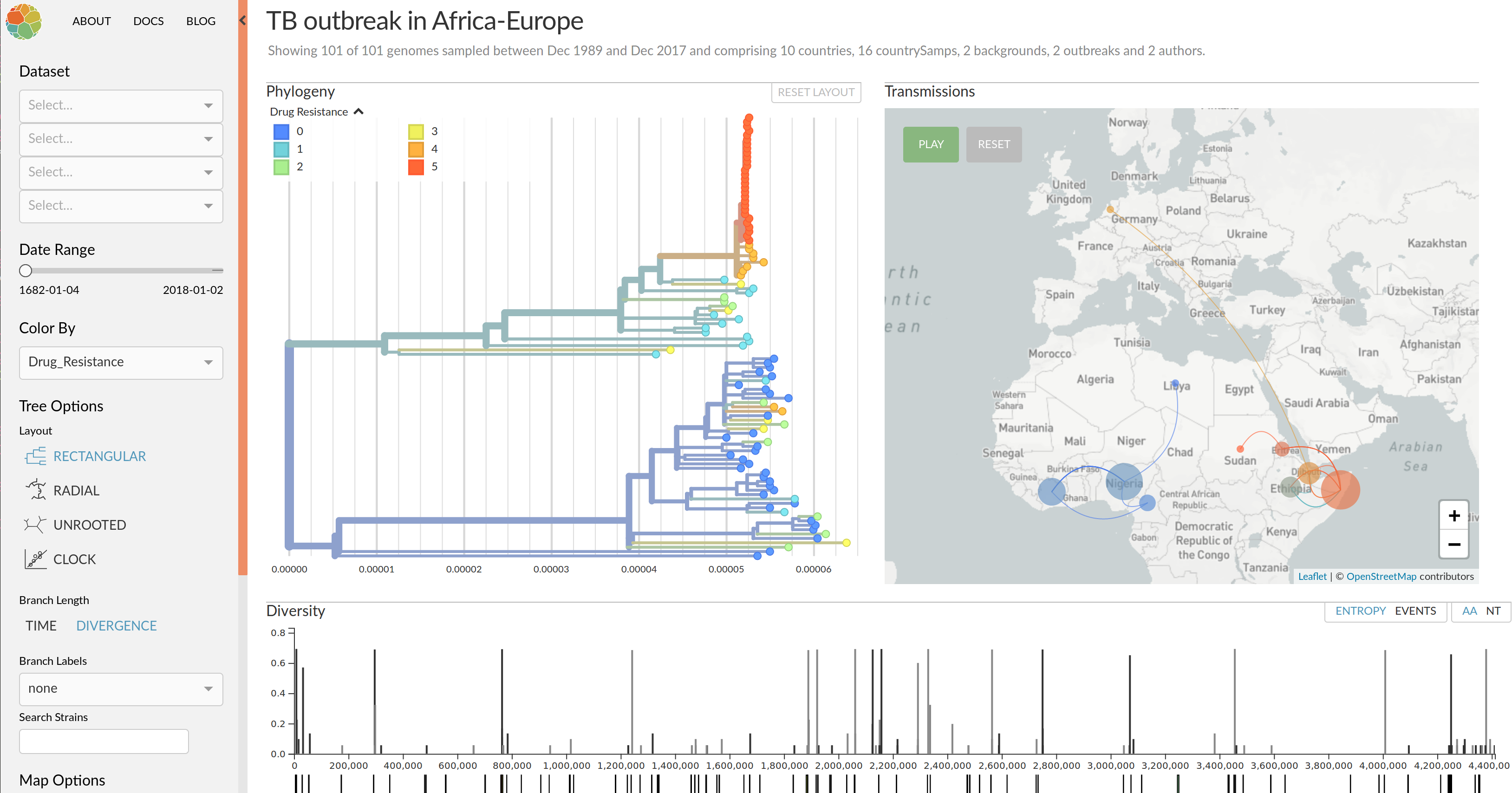
Visualization features of nextstrain
- Regular and time scaled phylogenies
- Mutations are mapped to the tree
- Filtering to time interval, region, country, authors, ...
- Zoom into clades
- Information on specific viruses
- Color by amino acid or nucleotide
- Frequency trajectories of clades and mutations
- Color by antigenic advance, predictive scores, etc
Phylodynamic analysis with nextstrain
- input: metadata (csv table) + sequences
- composable snakemake pipeline
- filtering (select relevant strains)
- alignment
- tree building (optionally time scaled trees)
- ancestral state reconstruction and phylogeography
- additional pathogen specific steps
- export to visualization
- runs in minutes to 1h
Web visualization with nextstrain
- fairly easy to set-up
- can be run locally (localhost)
- or be deployed on your own servers
- work in progress:
- flexible branding
- drag and drop features
- (better docs...)
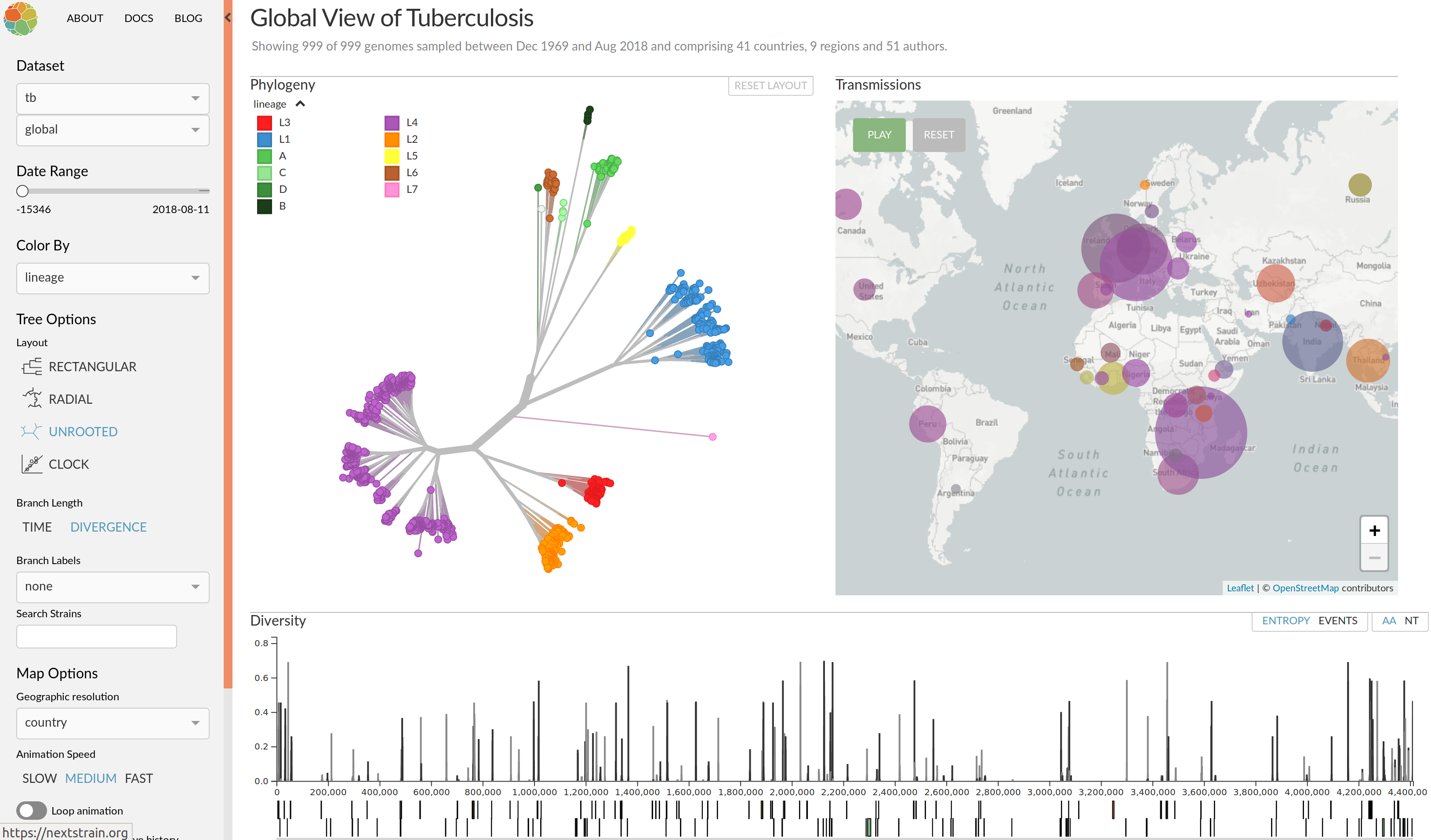

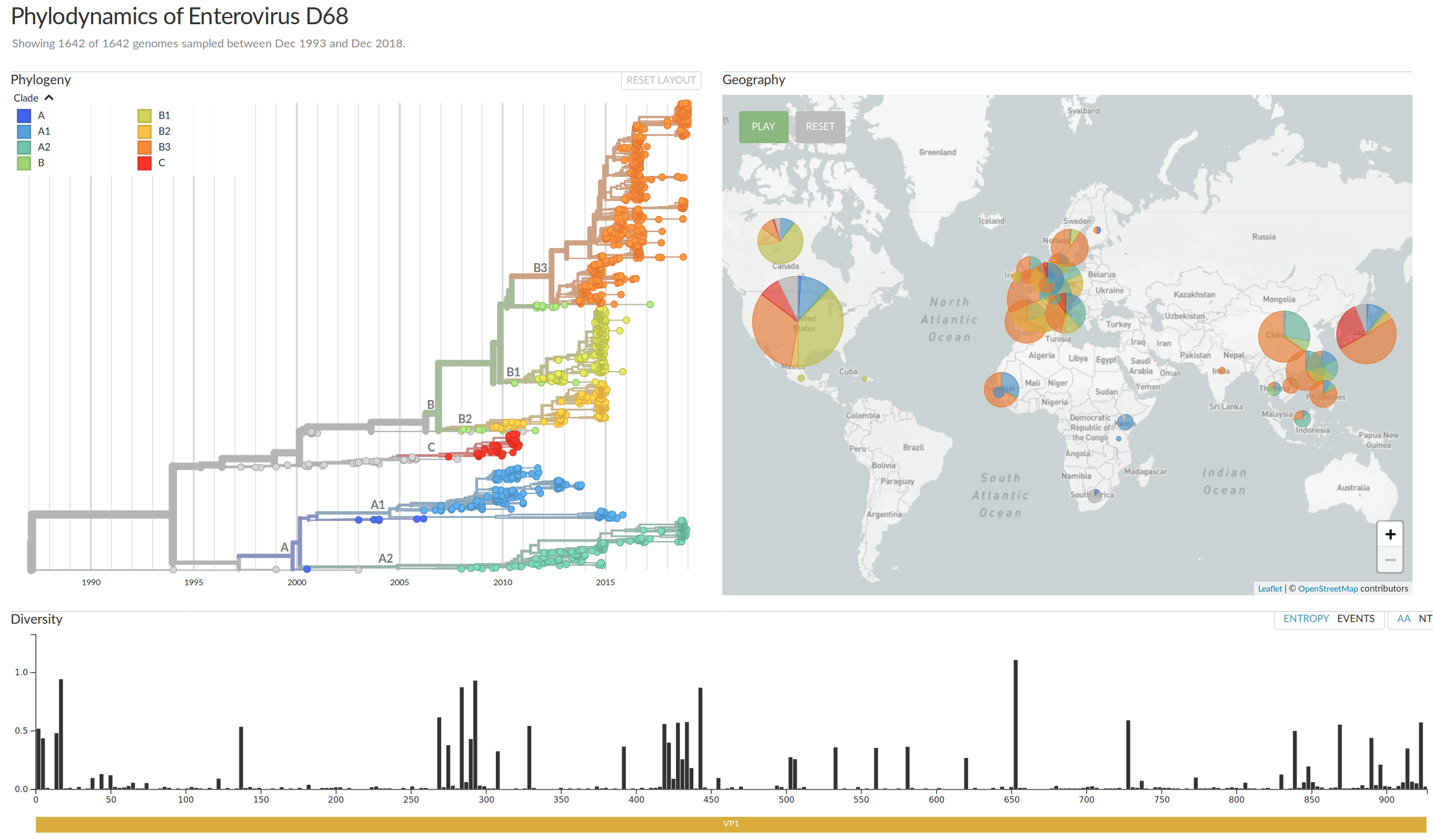
Enterovirus D68 -- with Robert Dyrdak, Emma Hodcroft & Jan Albert
- Non-polio enterovirus
- Almost everybody has antibodies against EV-D68
- Large outbreak in 2014 with severe neurological symptoms in
young children (acute flaccid myelitis) - Another outbreak in 2016
- Outbreaks tend to start in late summer/fall
- Several reports of EV-D68 outbreaks last fall
(201 AFM cases in the US in 2018)
EV-D68 whole genome deep sequencing project across Europe
Infections with multiple variants
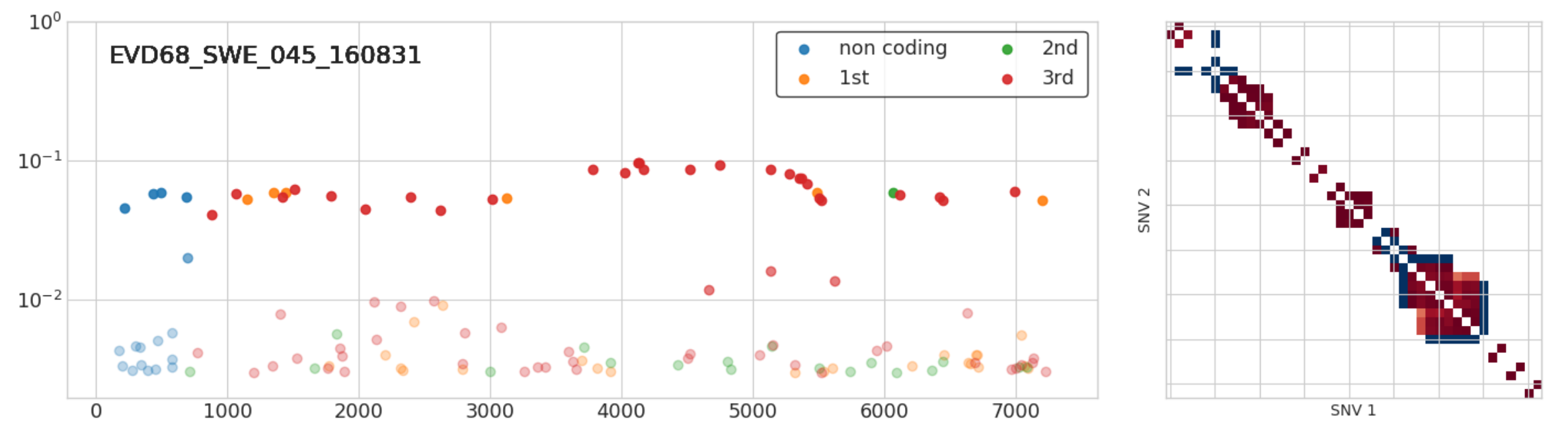
- A set of iSNVs at very similar frequencies in full linkage
- Suggest infection with two related variants
- 3 out of 50 samples: Implies high prevalence
Acknowledgments





- Trevor Bedford
- Colin Megill
- Pavel Sagulenko
- Sidney Bell
- James Hadfield
- Emma Hodcroft
- and others




Acknowledgments -- Enterovirus
- Robert Dyrdak
- Jan Albert
- Lina Thebo
- Emma Hodcroft
- Bert Niesters (Groningen)
- Randy Poelman (Groningen)
- Elke Wollants (Leuven)
- Adrian Egli (Basel)
- Andrés Antón Pagarolas (Barcelona)
