Repeatability and predictability in microbial evolution
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/201907_Nordita.html
- Repeatability in replicated experiments
- same exact mutations?
- same genes?
- same pathways?
- same phenotypes?
- Predictability
- future frequency trajectories of existing alleles?
- future mutations?
Colistin resistance evolution in Pseudomonas aeruginosa
- polymyxin, active against gram negatives
- interacts with outer membrane
- old antibiotic, discontinued because of nephrotoxicity
- today: last-resort antibiotic
- How fast?
- Which mutations?
- Which order?
- Genetic background?
Morbidostat by Toprak et al.
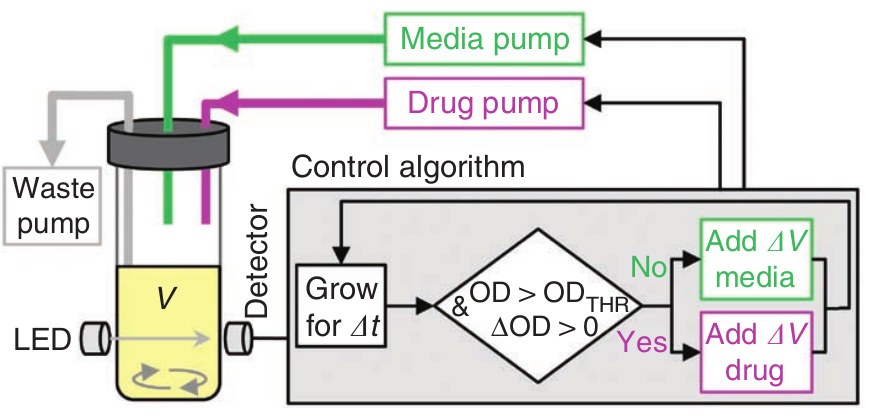
- Computer measures OD
- Controls pumps to add medium or AB
- Waste is removed
- Morbidostat→ growth rate is kept constant
- Chemostat → dilution is constant
- Turbidostat → OD is constant
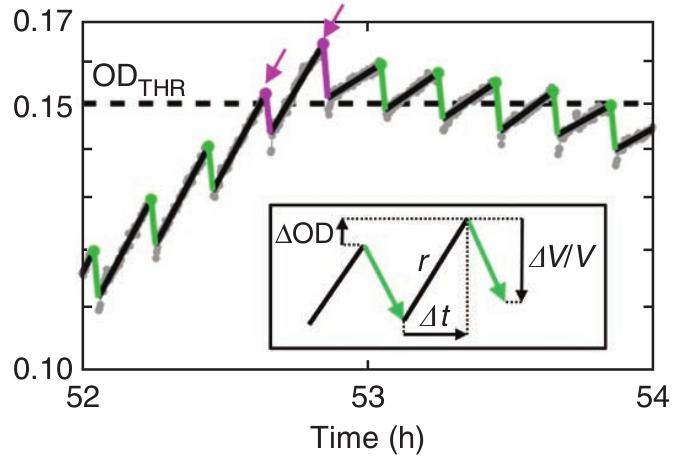
Our morbidostat

- more flexible software
- more compact design
- cheaper pumps and controllers


Colistin resistance emerges within 2 weeks
Mutation trajectories in strain PA77
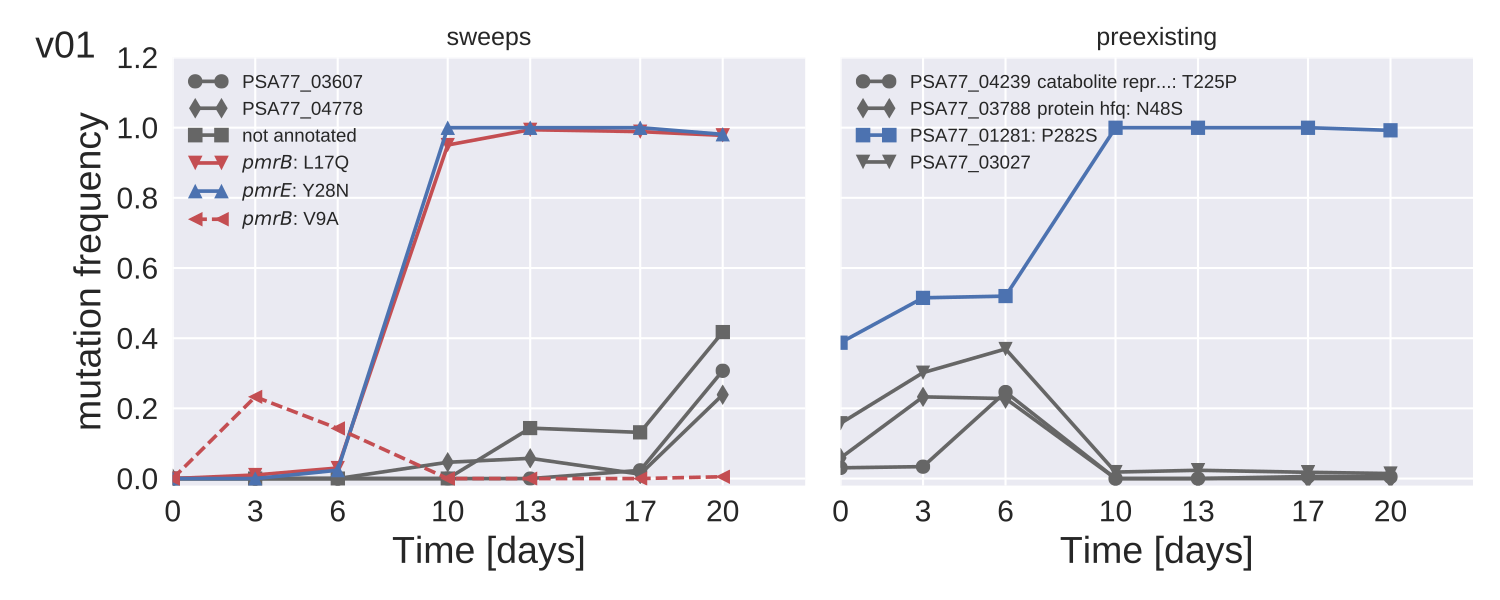
- Whole genome deep sequencing ($>200x$) with Illumina.
- Track rare mutations, no matter where they are
- Mutation frequencies to about 5% accuracy
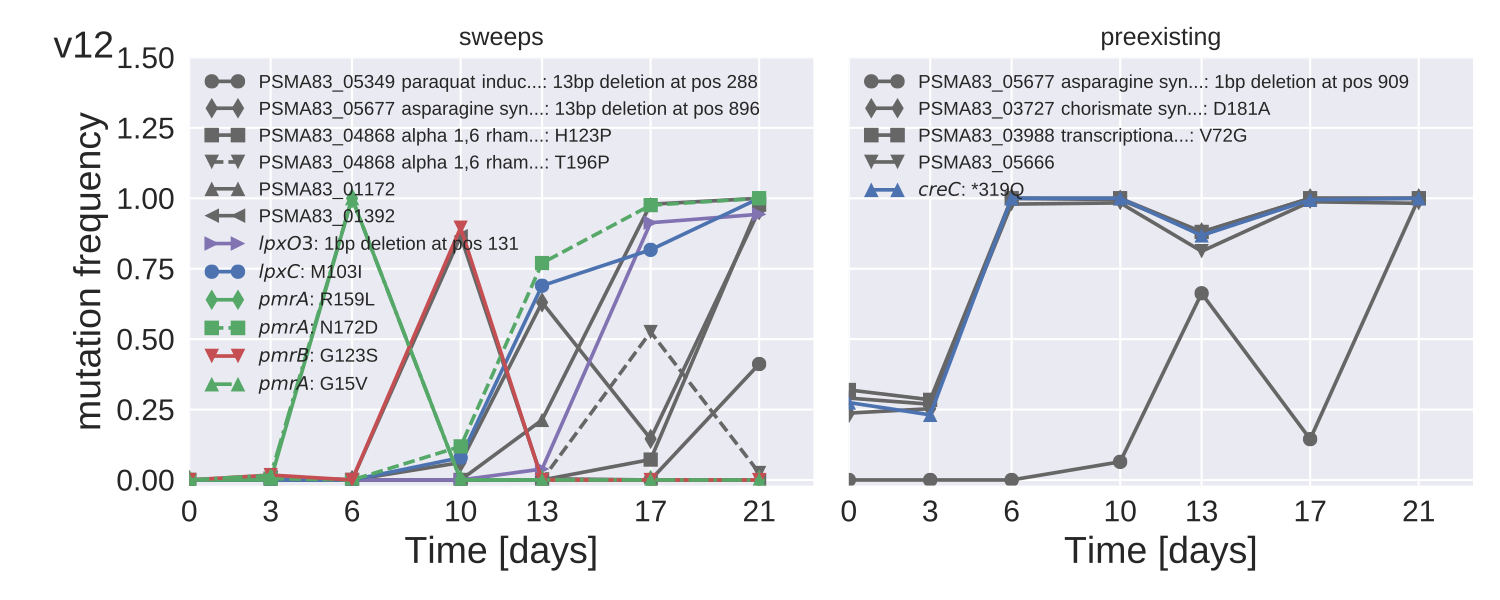
Mutation trajectories in strain PA83
Recurrent mutations PA77
| Gene | locus tag PAO1 | v01 | v02 | v03 | v04 | v05 | v05a | v08a | v10a | v11a |
| pmrB | PA4777 | V9A,L17Q | L90Q,E320K | V9A | P216Q | P254L | P169X,M292I | S257N | N41I,P169X | H261Y |
| pmrE | PA2043 | Y28N | Y28C | Y28N | Y28C | Y28N | Y28C | Y28C | Y28N | Y28N |
| lptD | PA3559 | Y803X | L538R |
- pmrE: Most PA strains are 28C → reversion
- pmrB: Many mutations that constitutively activate the gene
→ canonical colistin resistance gene - lptD: code for outer membrane protein, LPS transport.
→ has been associated with colistin resistance in Acinetobacter
Recurrent mutations in PA83
| locus tag PAO1 | v02 | v03 | v05 | v06 | v08 | v11 | v12 | v14 | v15 | |
| lpxC | PA4406 | P101S | V222A,S106G | V222A | V164G,A107T | A107T,G21W,F176S | A107T,I131F | M103I | D232E,D232G,V217F,V217A | V222A,S106G |
| pmrB | PA4777 | L96R | L171P | L87P | F51L | S8P,E320K | V9A | G123S | E320K,A248T,L167P | R259H,V361M |
| putative tranferase | PA3853 | C226G | Y3C,G62S | V34A,Y155C | C226G | R60C,Y216C,E185G | C226G | V122A,E185G | ||
| asparagine synthetase | L365P | frameshift | L425P | G32S | frameshift | W153* | L365P,W153*,V286M | |||
| migA | PA0705 | H219P | C25R,N27S | D106G | Q191R,V22A | T196P,H123P | H219P | A168T | ||
| mutS | PA3620 | T51P | T51P | T51P | T51P | T51P | T51P,T287P | |||
| lpxO2 | PA0936 | D163A | D163N | W209* | D163A | frameshift | in-frame deletion | |||
| pmrA | PA4776 | L11Q | L11P | R159L,G15V,N172D | ||||||
| cupB5 | PA4082 | G260X,R26C | P139P | |||||||
| pdtA | PA0690 | A3885V,A3885A | G1527X | |||||||
| morA | PA4601 | R1199H | G143D | |||||||
| lpxA | PA3644 | R96S | R191C | |||||||
| priA | PA5050 | L38L | R689R | |||||||
| traN | W773* | G912D | ||||||||
| wbpM | PA3141 | E273K | E273G | |||||||
| mscL | PA4614 | V86I | S35P |
- lpxC: lipid A biosynthesis
- pmrB: Many mutations that constitutively activate the gene
→ canonical colistin resistance gene - lptD: code for outer membrane protein, LPS transport.
→ has been associated with colistin resistance in Acinetobacter
Mutations in pmrAB
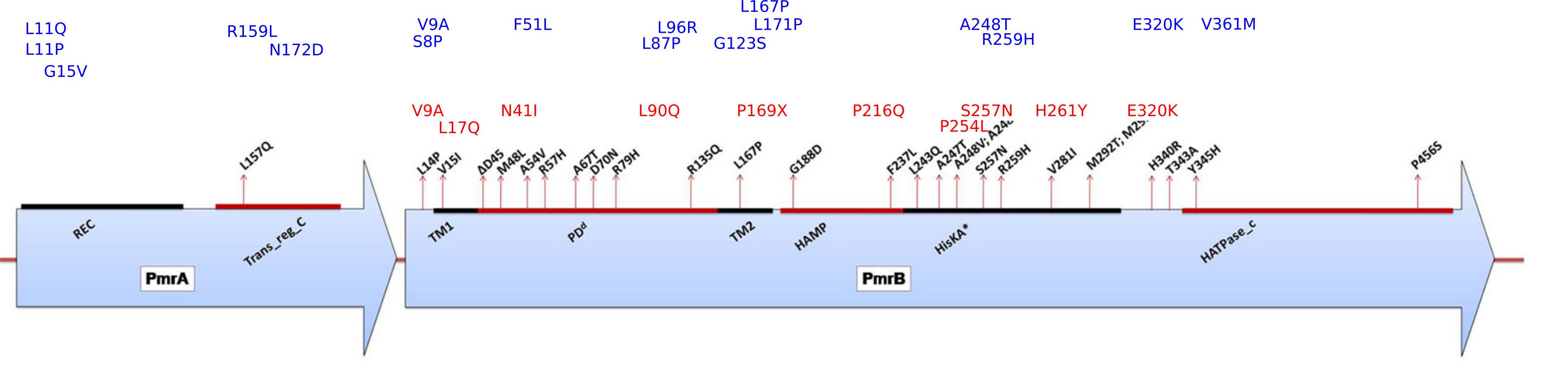 Previously observed and new mutations in pmrAB (blue: PA83, red: PA77)
Olaitan et al. Front. Micro., 2014
Previously observed and new mutations in pmrAB (blue: PA83, red: PA77)
Olaitan et al. Front. Micro., 2014
Evolution of HIV
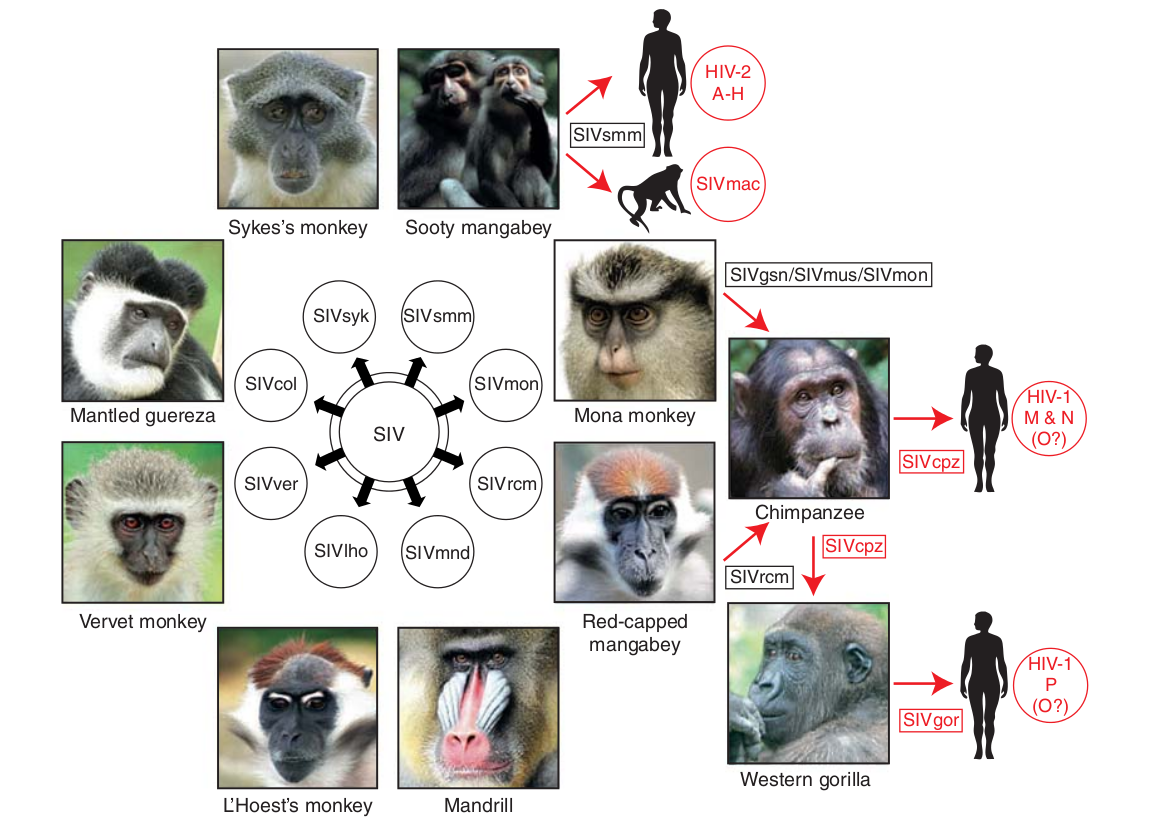
- Chimp → human transmission around 1900 gave rise to HIV-1 group M
- ~100 million infected people since
- subtypes differ at 10-20% of their genome
- HIV-1 evolves ~0.1% per year
HIV infection
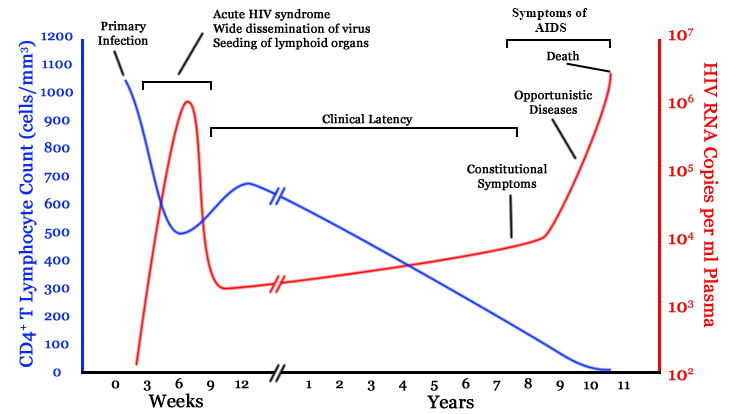
- $10^8$ cells are infected every day
- the virus repeatedly escapes immune recognition
- integrates into T-cells as
latent provirus
HIV-1 evolution within one individual
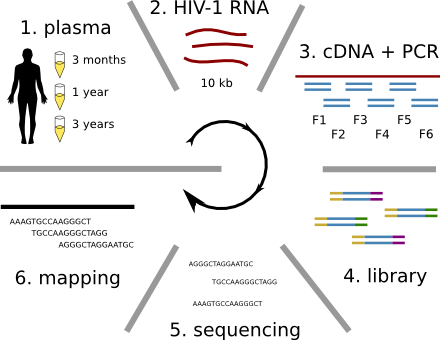

Population sequencing to track all mutations above 1%
Minor diversity accumulation is predictable
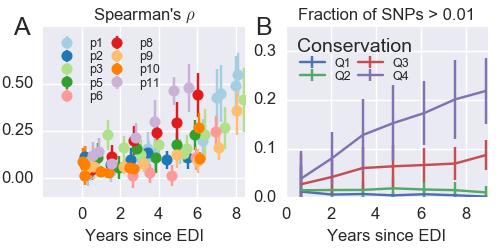
Divergence at increasingly conserved positions
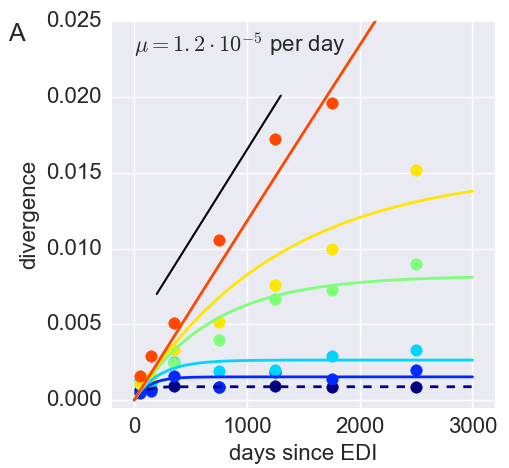
- Six categories from high to low conservation
- mutation away from preferred state with rate $\mu$
- selection against non-preferred state with strength $s$
- variant frequency dynamics: $\frac{d x}{dt} = \mu -s x $
- equilibrium frequency: $\bar{x} = \mu/s $
- fitness cost: $s = \mu/\bar{x}$
- Fit model of minor variation to categories of conservation
- $\Rightarrow$ harmonic average fitness cost in category
Fitness landscape of HIV-1
Zanini et al, biorxiv, 2017Selection on RNA structures and regulatory sites
Zanini et al, biorxiv, 2017


- Influenza viruses evolve to avoid human immunity
- Vaccines need frequent updates

Fitness variation in rapidly adapting populations
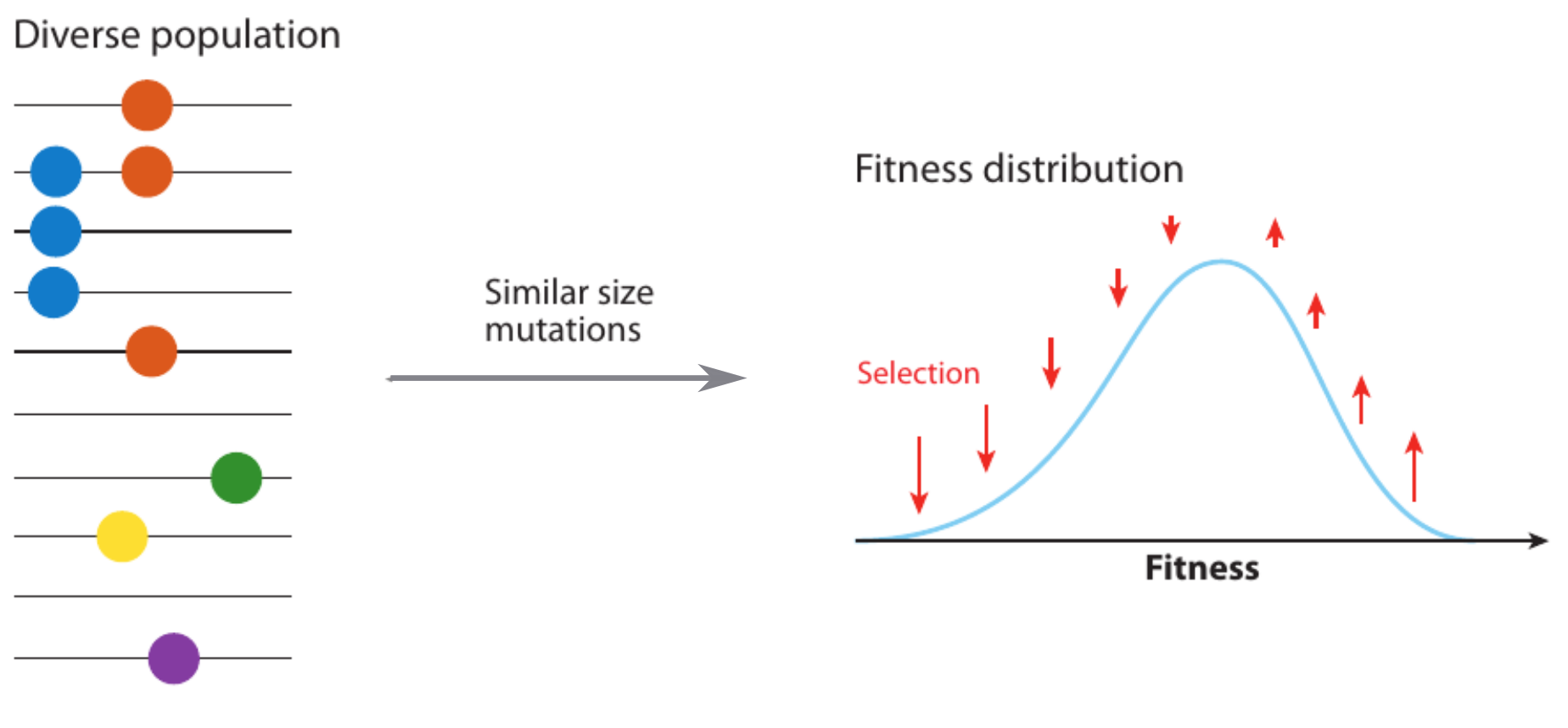
- Speed of adaptation is logarithmic in population size
- Environment (fitness landscape), not mutation supply, determines adaptation
- Different models have universal emerging properties
Neutral/Kingman coalescent
strong selection
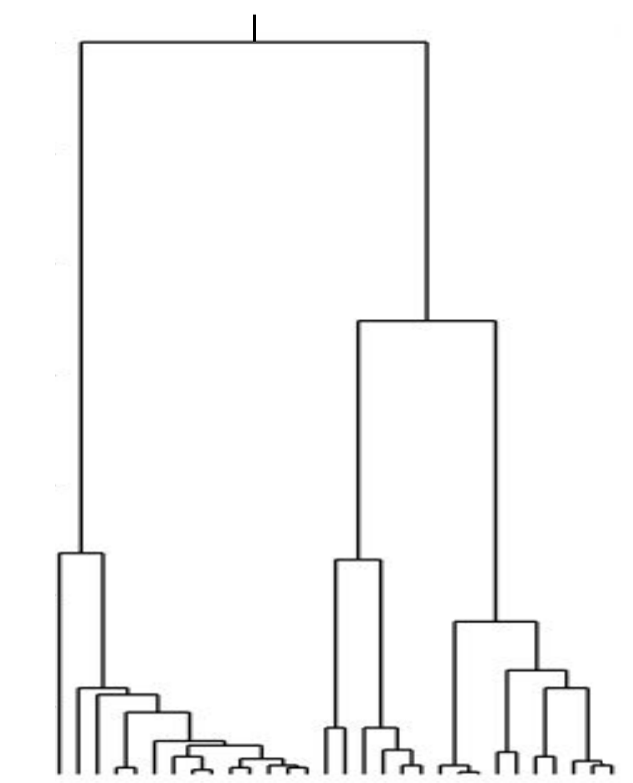

Bolthausen-Sznitman Coalescent
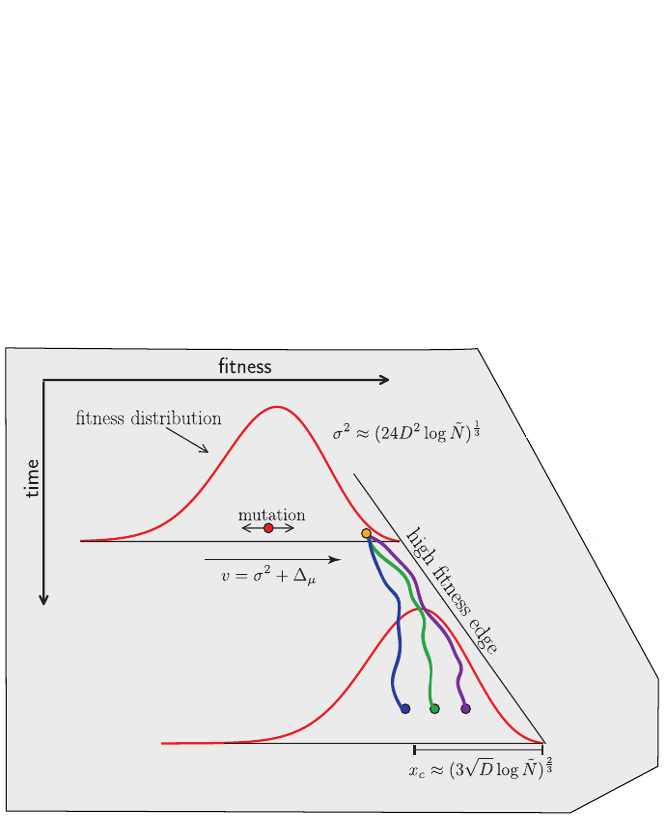
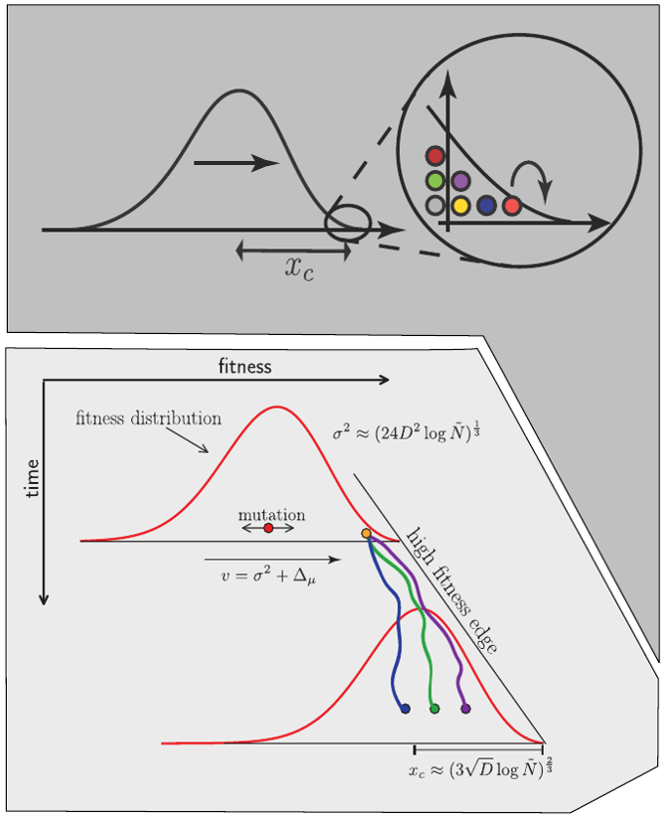
Bursts in a tree ↔ high fitness genotypes
Can we read fitness of a tree?
Predicting evolution

Given the branching pattern:
- can we predict fitness?
- pick the closest relative of the future?
Fitness inference from trees
$$P(\mathbf{x}|T) = \frac{1}{Z(T)} p_0(x_0) \prod_{i=0}^{n_{int}} g(x_{i_1}, t_{i_1}| x_i, t_i)g(x_{i_2}, t_{i_2}| x_i, t_i)$$
RN, Russell, Shraiman, eLife, 2014
Prediction of the dominating H3N2 influenza strain
- no influenza specific input
- how can the model be improved? (see model by Luksza & Laessig)
- what other context might this apply?
Summary
- Colistin resistance evolution predictable at the gene and pathway level
- Intra-host HIV evolution is governed by a universal fitness landscape, modulated by host-specific immune response
- Reversion and and predictable diversity patterns
- Landscape of fitness costs can be estimated from intra-host diversity
- Tree shape contains fitness information -- estimate derivatives from a snapshot
Thank you for your attention!
Acknowledgements -- Colistin
- Bianca Regenbogen, now Uni Hohenheim
- Silke Peter, UKT Tübingen
- Matthias Willmann, UKT Tübingen


Acknowledgments -- HIV
- Fabio Zanini
- Jan Albert
- Johanna Brodin
- Christa Lanz
- Göran Bratt
- Lina Thebo
- Vadim Puller



Influenza and Theory acknowledgments




- Boris Shraiman
- Colin Russell
- Trevor Bedford
- Oskar Hallatschek


