Within-host evolution of HIV
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/201910_Amsterdam.html
The people who made this happen...
- Fabio Zanini
- Jan Albert
- Johanna Brodin
- Christa Lanz
- Göran Bratt
- Lina Thebo
- Vadim Puller
- The participants of this study!



Evolution of HIV
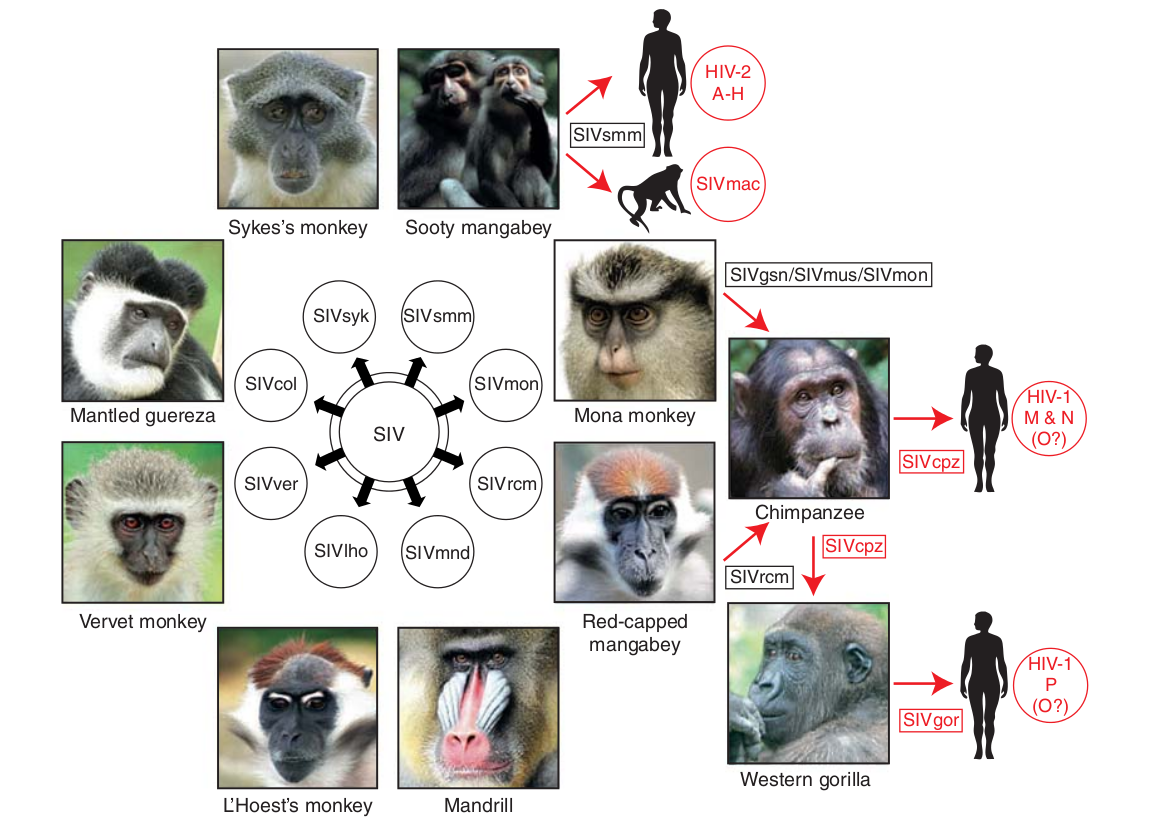
- Chimp → human transmission around 1900 gave rise to HIV-1 group M
- ~100 million infected people since
- subtypes differ at 10-20% of their genome
- HIV-1 evolves ~0.1% per year
HIV infection
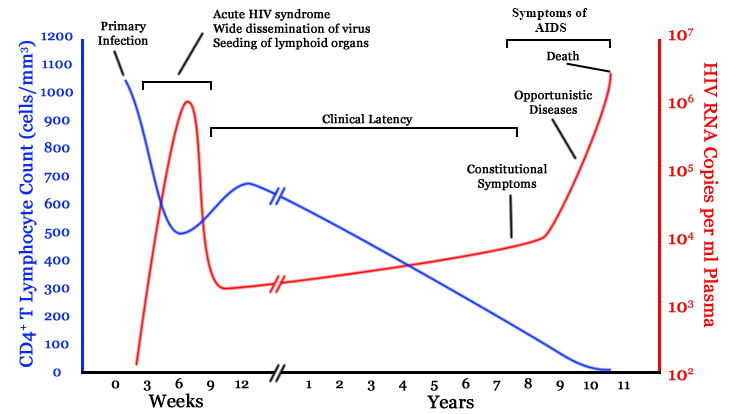
- $10^8$ cells are infected every day
- the virus repeatedly escapes immune recognition
- integrates into T-cells as
latent provirus
HIV-1 evolution within individuals

Untreated early to chronic infection:
Virus-immune system coevolution
Population sequencing to track all mutations above 1%
Evolution and diversity across the genome
- envelope changes fastest, enzymes slowest
- identical rate of synonymous evolution
- diversity saturates where evolution is fast
- synonymous mutations stay at low frequency
Approximately neutral divergence at silent mutations
In vivo mutation rate estimates
Frequent reversion towards the center of HIV phylogeny
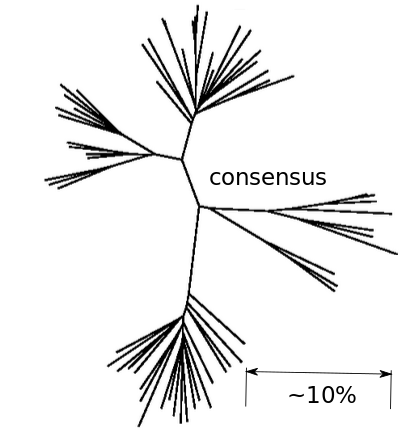
- Almost one third of all mutations are reversions (~5% expected by chance)
- Reversions accumulate slowly throughout chronic infection
- Reversion can explain much of the intra/inter-host evolutionary rate mismatch
Divergence at increasingly conserved positions
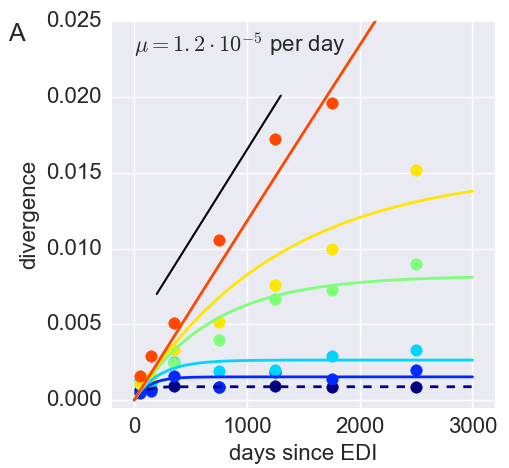
- Six categories from high to low conservation
- mutation away from preferred state with rate $\mu$
- selection against non-preferred state with strength $s$
- variant frequency dynamics: $\frac{d x}{dt} = \mu -s x $
- equilibrium frequency: $\bar{x} = \mu/s $
- fitness cost: $s = \mu/\bar{x}$
- Fit model of minor variation to categories of conservation
- $\Rightarrow$ harmonic average fitness cost in category
Fitness landscape of HIV-1
Zanini et al, Virus Evolution, 2017Suppressive therapy and the latent reservoir:
Where is HIV hiding?
The latent proviral DNA reservoir
- Viruses integrate but don't express → pro-viral DNA reservoir
- Therapy fully suppresses virus replication, but interruption results in rebound
- When is the reservoir established? How is it maintained?
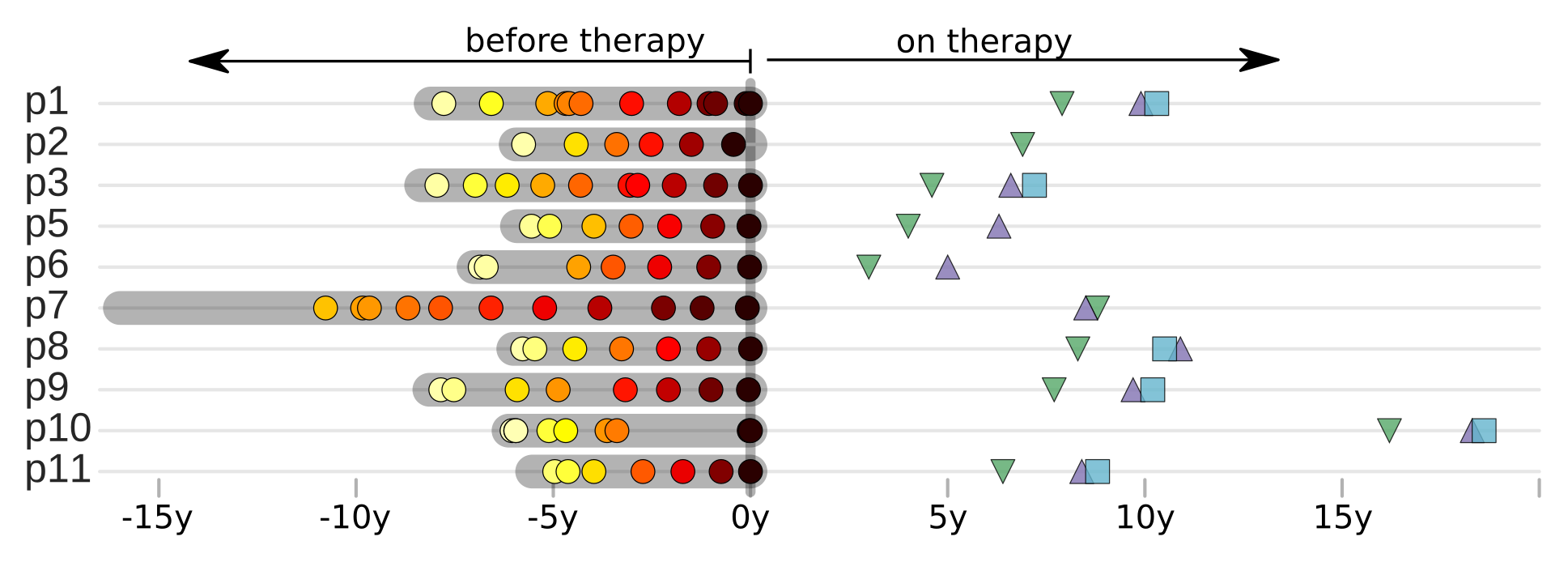
p17 sequencing from PBMCs on fully suppressive therapy
 Brodin et al, eLife, 2016
Brodin et al, eLife, 2016
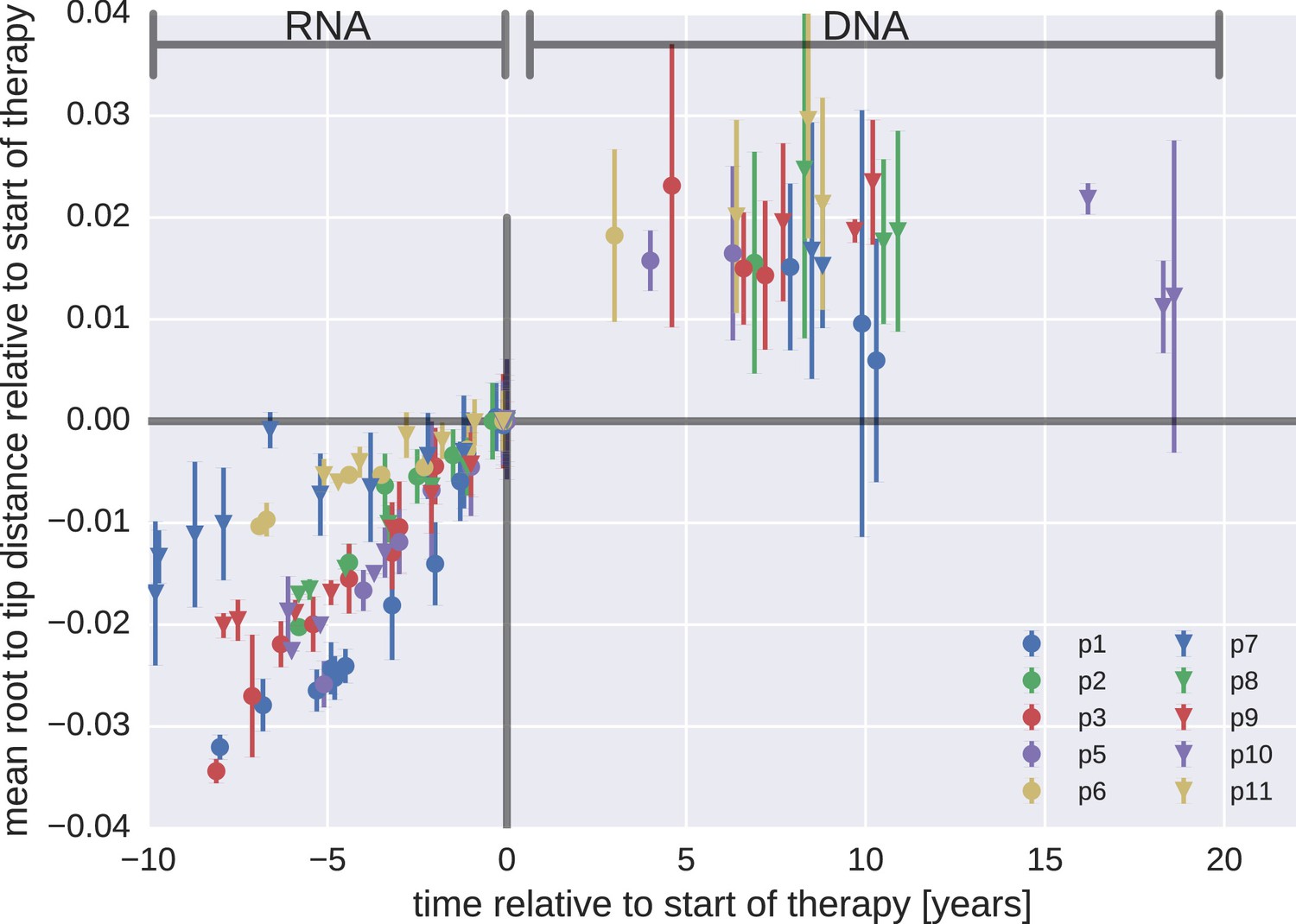
Proviral DNA mostly identical to pre-therapy RNA
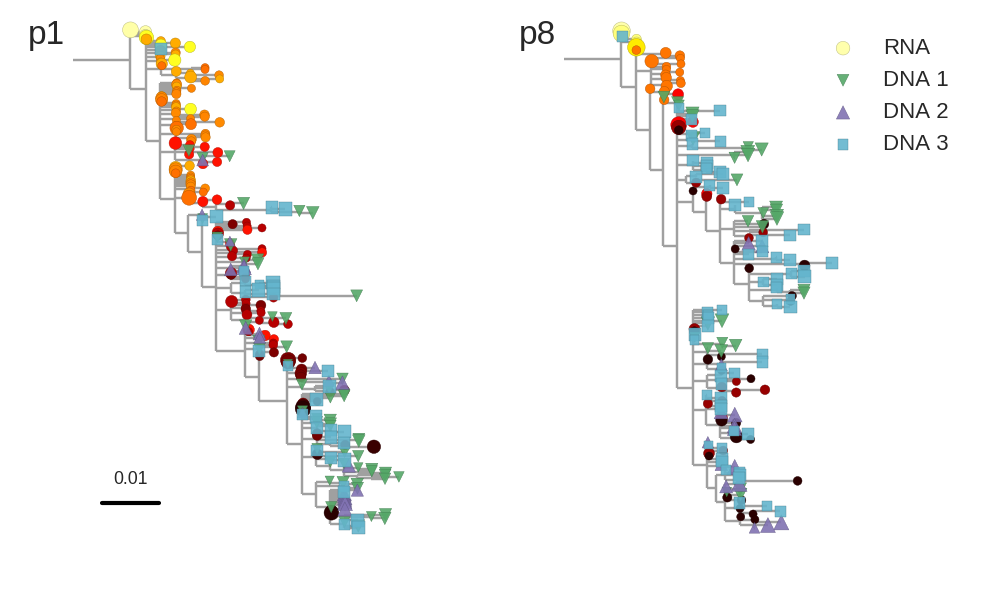
No evidence of ongoing evolution
Most proviral DNA originates from shortly after start of therapy
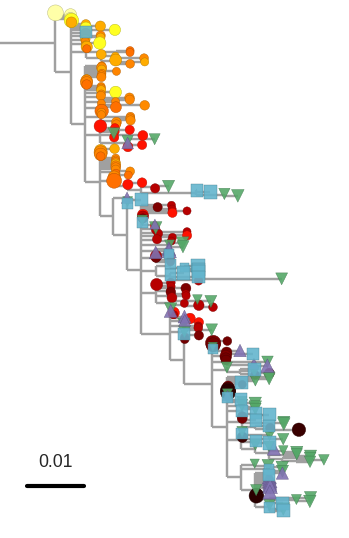
- latent HIV → barcode of a T-cell lineage
- all latent integrated virus derives from late infection
Summary
- Continuous reversion to ancestral states throughout chronic infection
- Minor diversity allows fitness cost estimates at every position in the genome
- The latent reservoir is a snapshot of the pre-treatment population
- No evidence of ongoing replication
- No clear trend of diversity loss on treatment
- Proviral DNA can serve as a T-cell lineage marker

Acknowledgments
- Fabio Zanini
- Jan Albert
- Johanna Brodin
- Christa Lanz
- Göran Bratt
- Lina Thebo
- Vadim Puller



No evidence of ongoing evolution -- hypermutated provirus
 Brodin et al, eLife, 2016
Brodin et al, eLife, 2016
Hypermutated and intact proviral p17 sequences
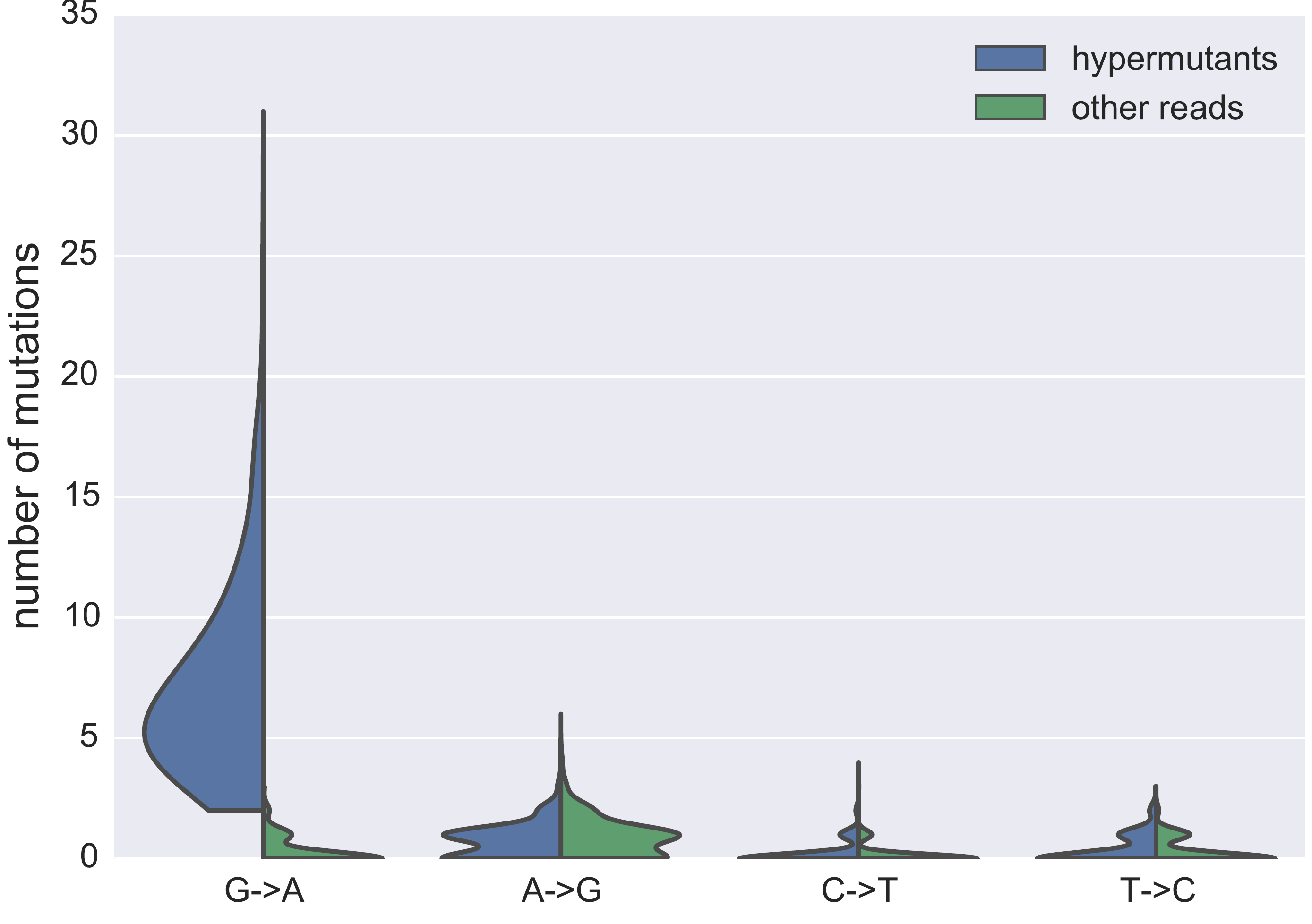
- Between 0 and 43% hypermutants, median 13%
- Between 3 and 43 unique sequences >1% read frequency, median 25
- Majority of hypermutant sequences have stop codons