Outbreak of Novel Coronavirus in Wuhan
Background, Spread, and Modelling
date: 2020-01-30
Richard Neher
Biozentrum & SIB, University of Basel
slides at neherlab.org/202001_nCov_en.html

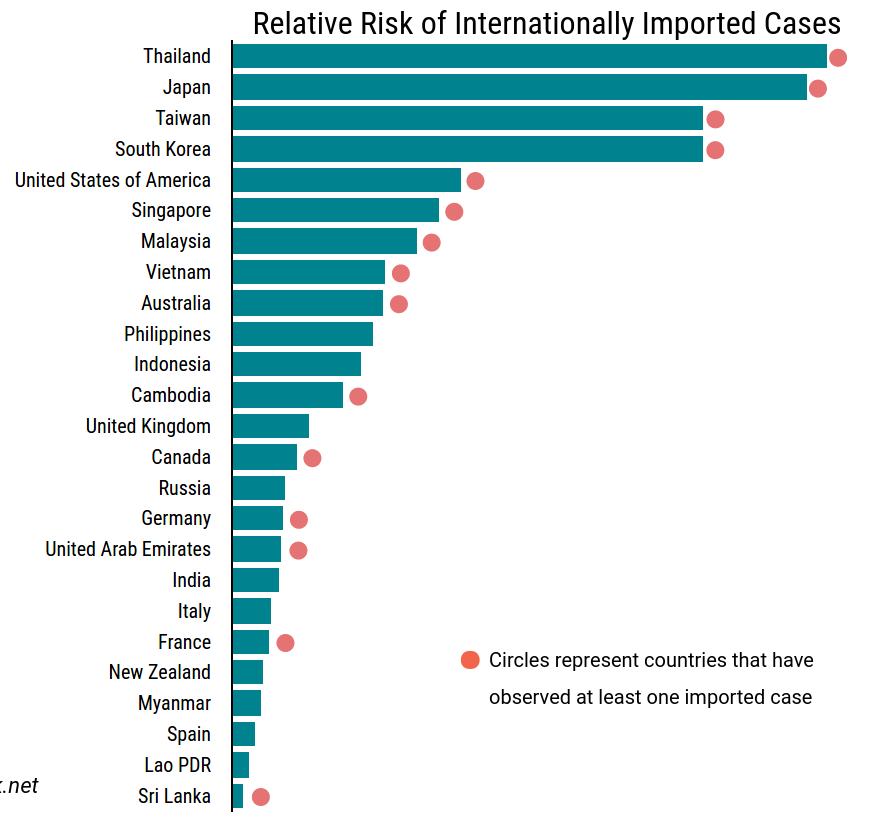
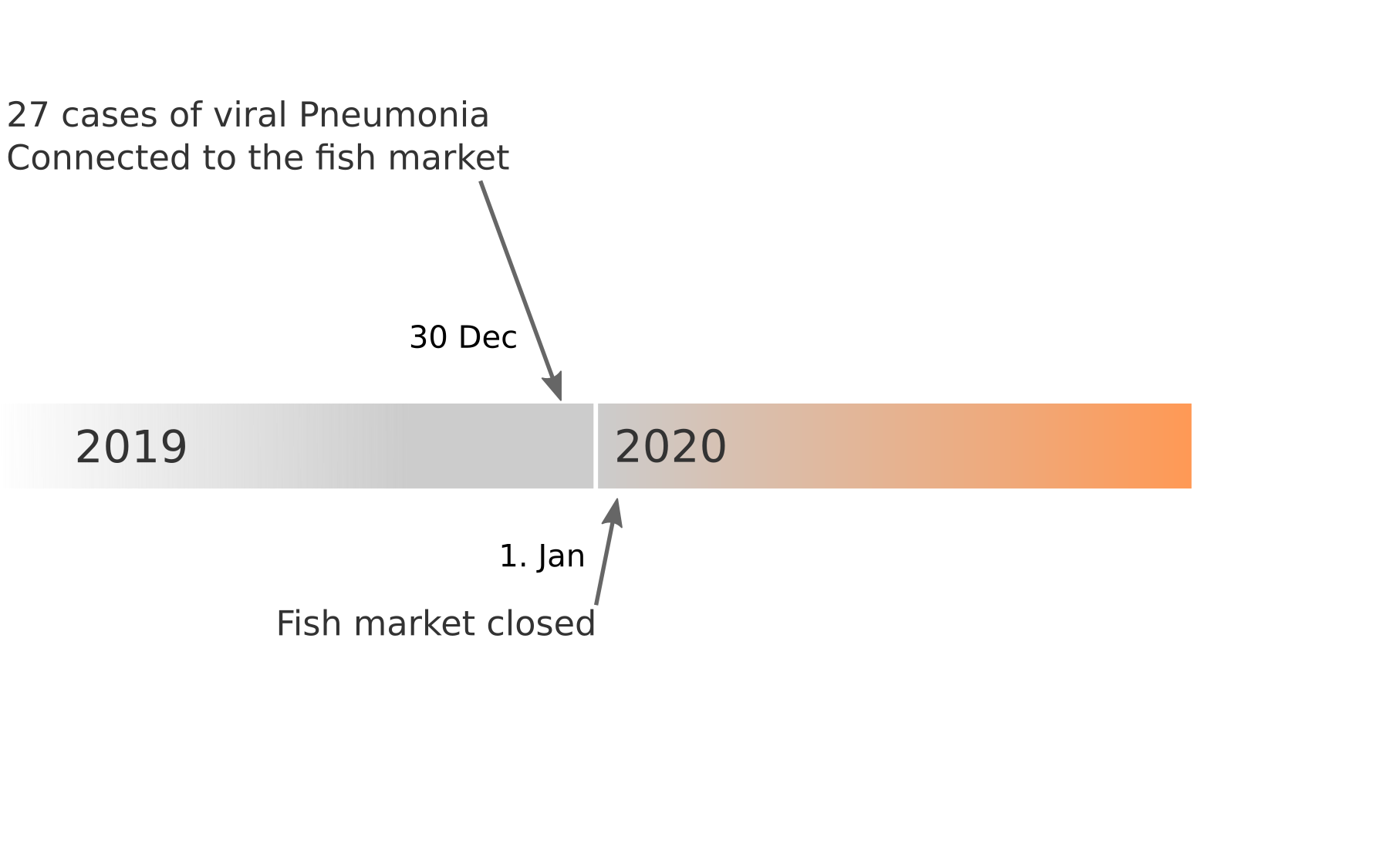 Data summarized by Ian MacKay
Data summarized by Ian MacKay

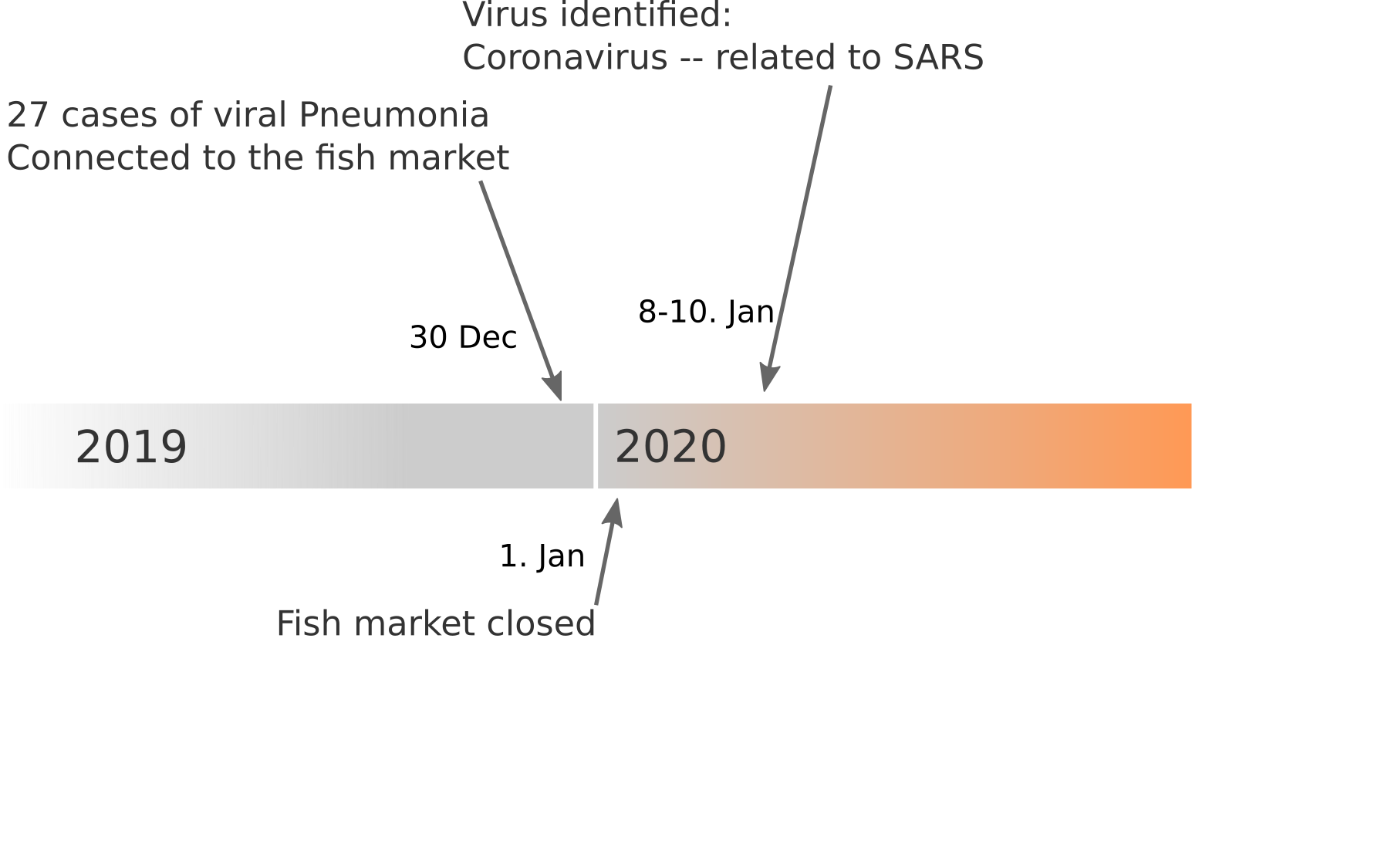 Data summarized by Ian MacKay
Data summarized by Ian MacKay

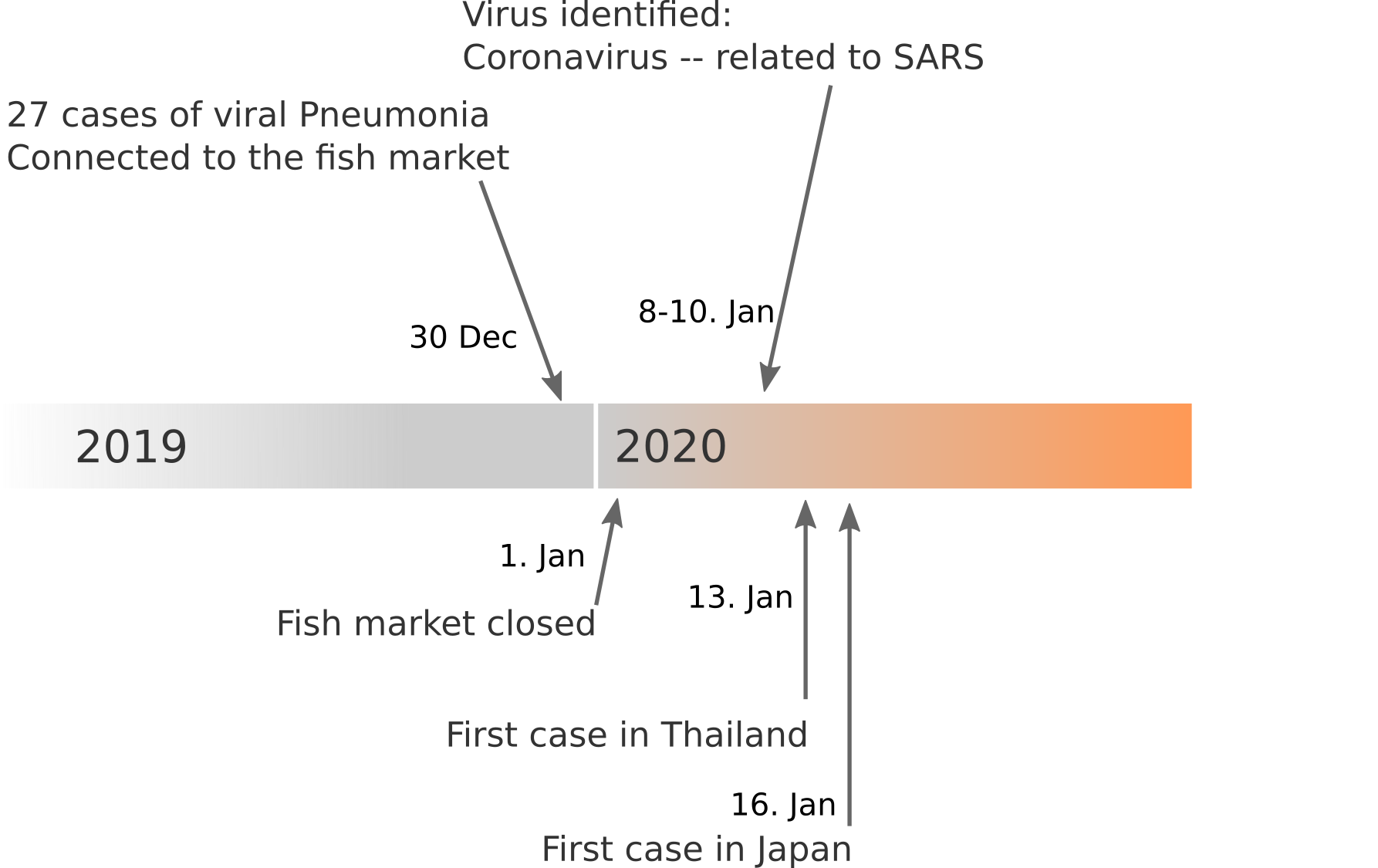 Data summarized by Ian MacKay
Data summarized by Ian MacKay

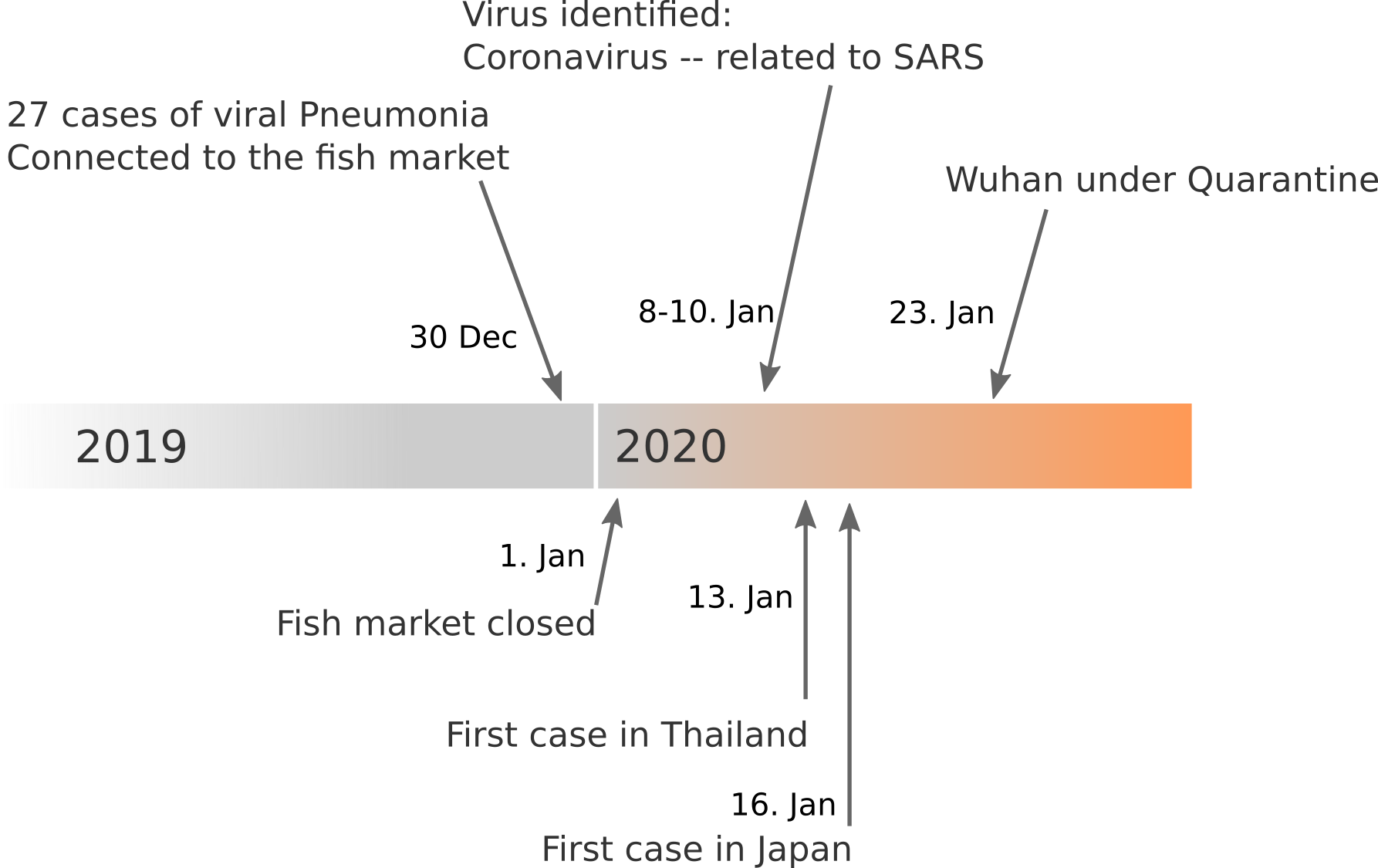 Data summarized by Ian MacKay
Data summarized by Ian MacKay

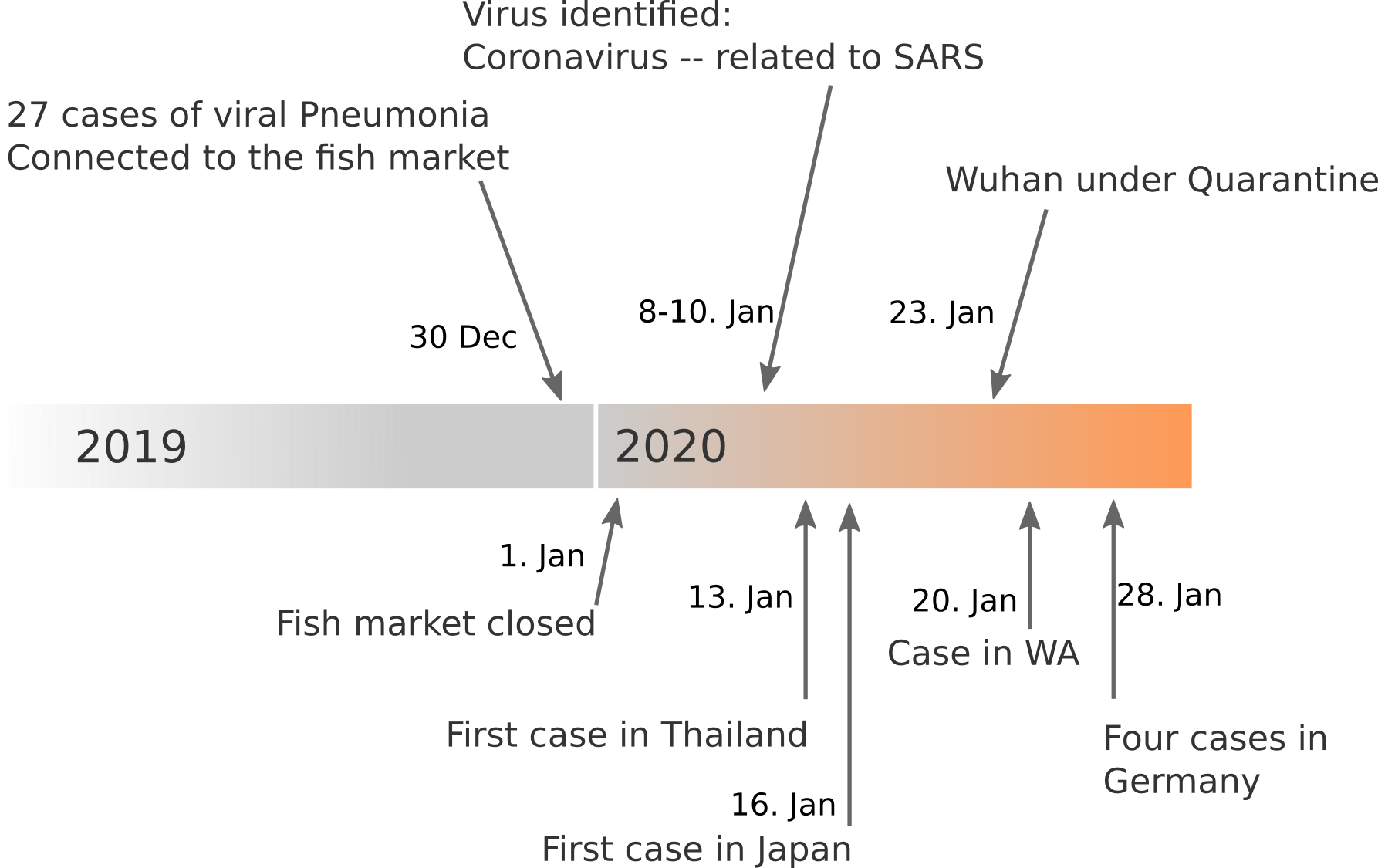 Data summarized by Ian MacKay
Data summarized by Ian MacKay
The first 41 cases from Wuhan (only hospitalized cases)


- Long incubation period makes prediction difficult.
- How many cases show any symptoms at all is uncertain.
Transmission

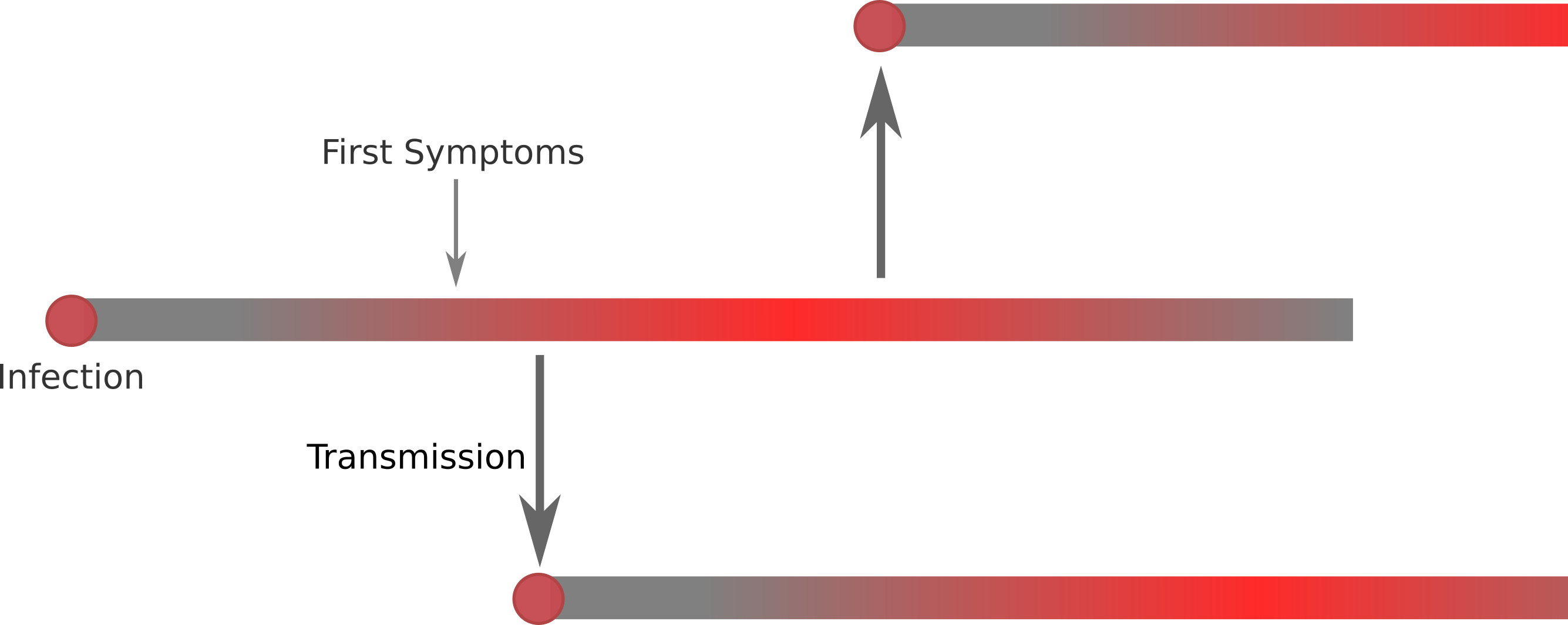
- The viruses is transmitted primarily by droplet infection.
- Whether someone is contagious before symptoms appear is unclear.
- Whether asymptomatic people can transmit is also unknown.
Transmission chains and unobserved cases




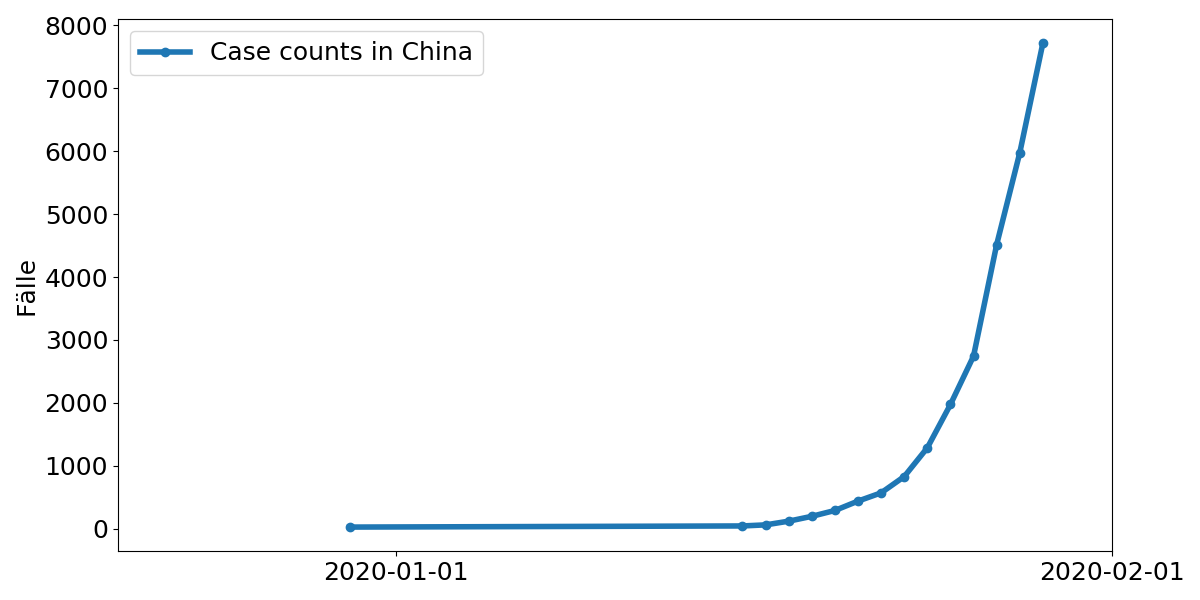
How fast does the virus spread?

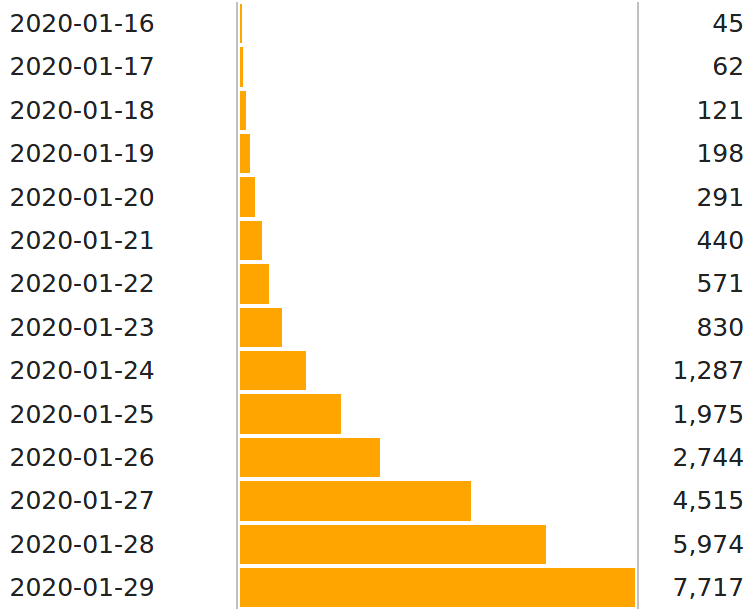
- Since the virus was identified, more and more tests are being done.
- The dynamics of the case numbers are therefore hard to interpret.
- How can we estimate dispersion more accurately?
How do we estimate the total number of infections?

- Outside China, cases are easier to find
- Infected person leaves China within a week: 1/1000
100 Exports → 10,0000 cases - Many cases are mild, the cases in Bavaria are also mild
- Rough estimation -- but plausible


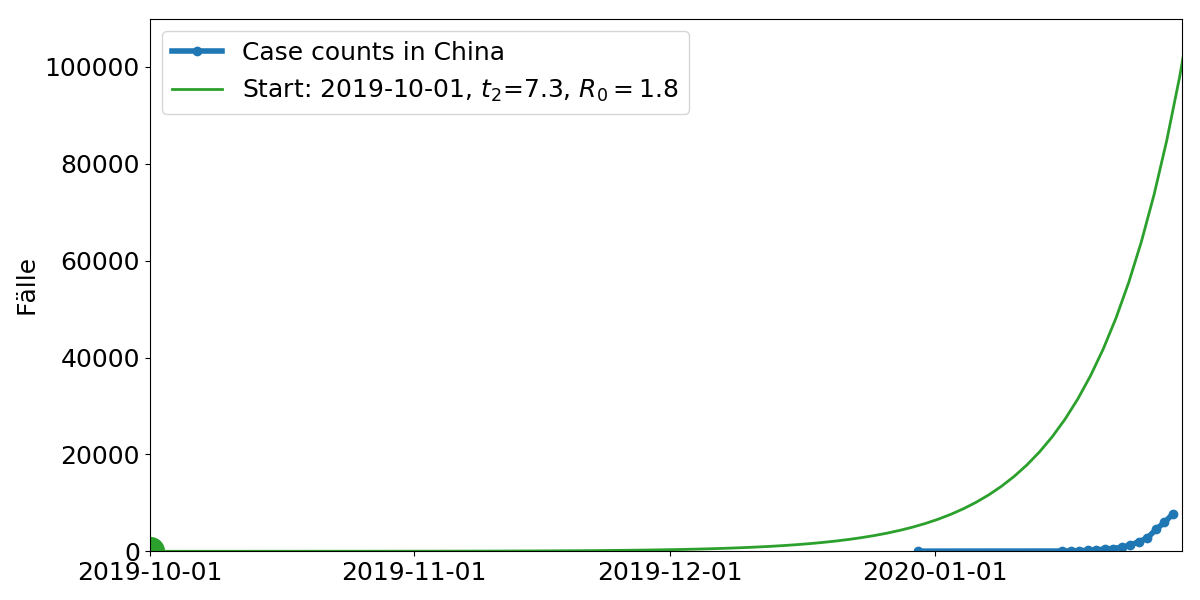
Model parameters: Start time, doubling time $t_2$. $R_0$ is the average number of subsequent cases

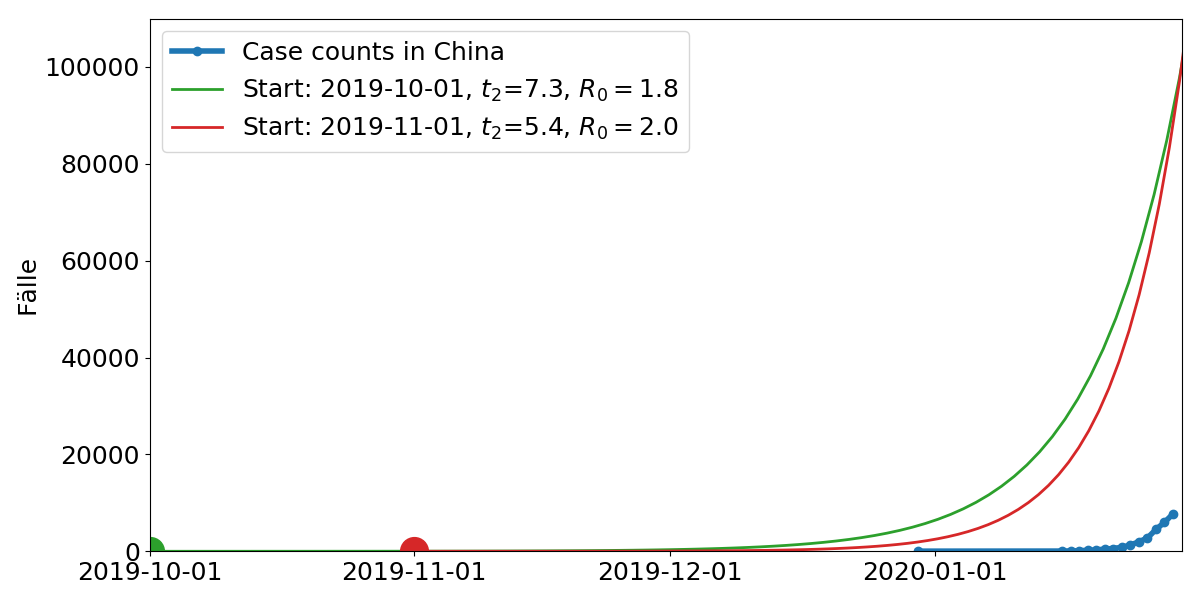
Model parameters: Start time, doubling time $t_2$. $R_0$ is the average number of subsequent cases

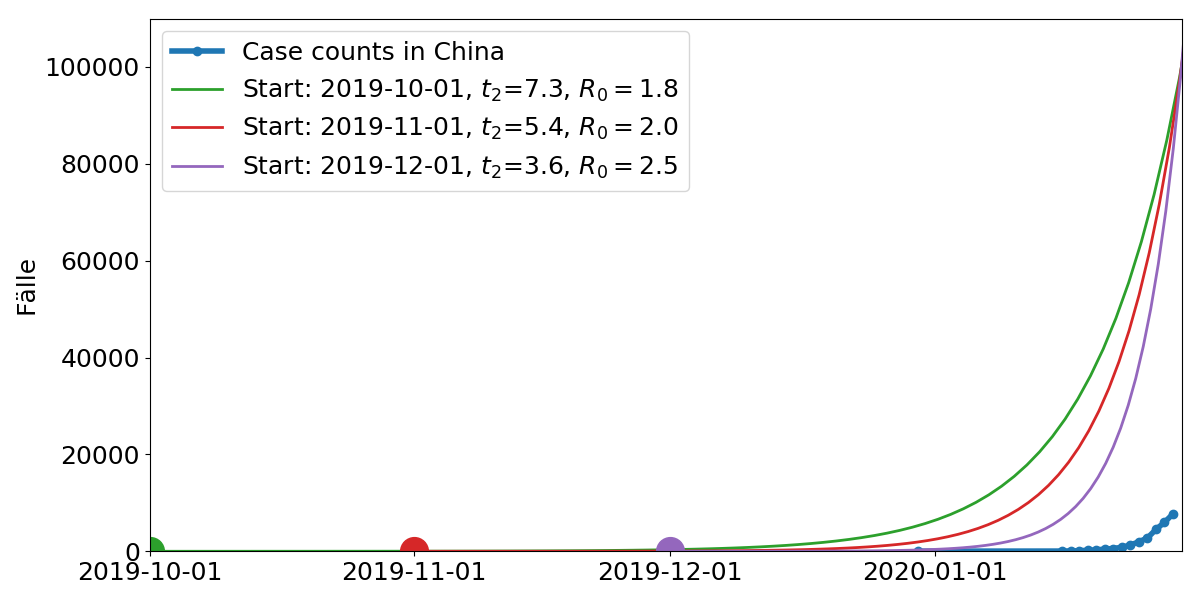
Model parameters: Start time, doubling time $t_2$. $R_0$ is the average number of subsequent cases

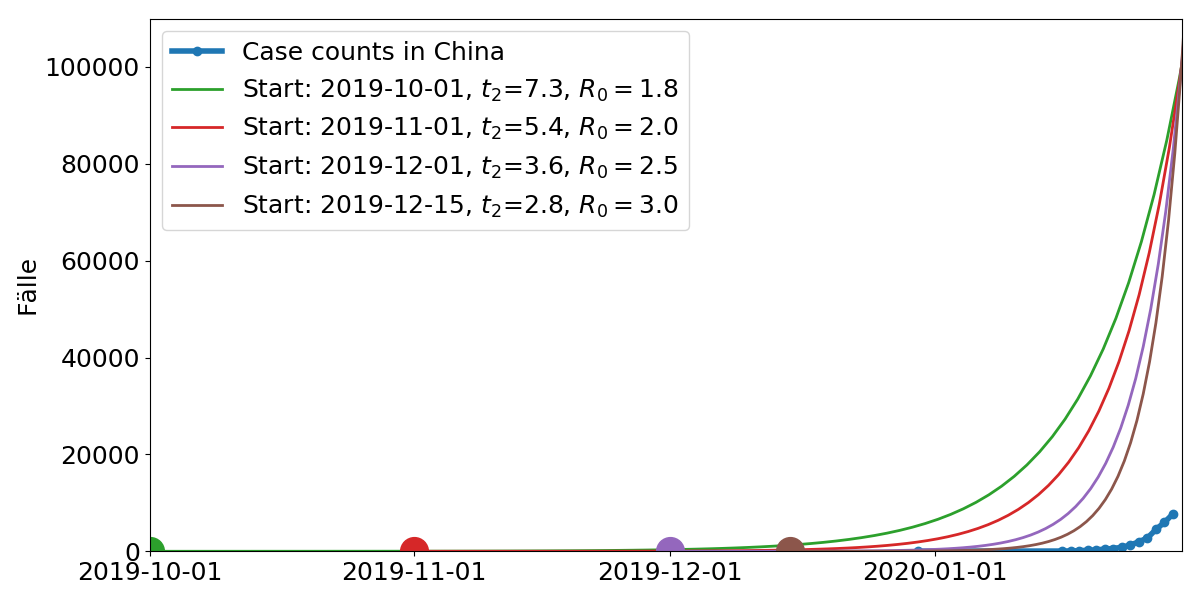
Model parameters: Start time, doubling time $t_2$. $R_0$ is the average number of subsequent cases

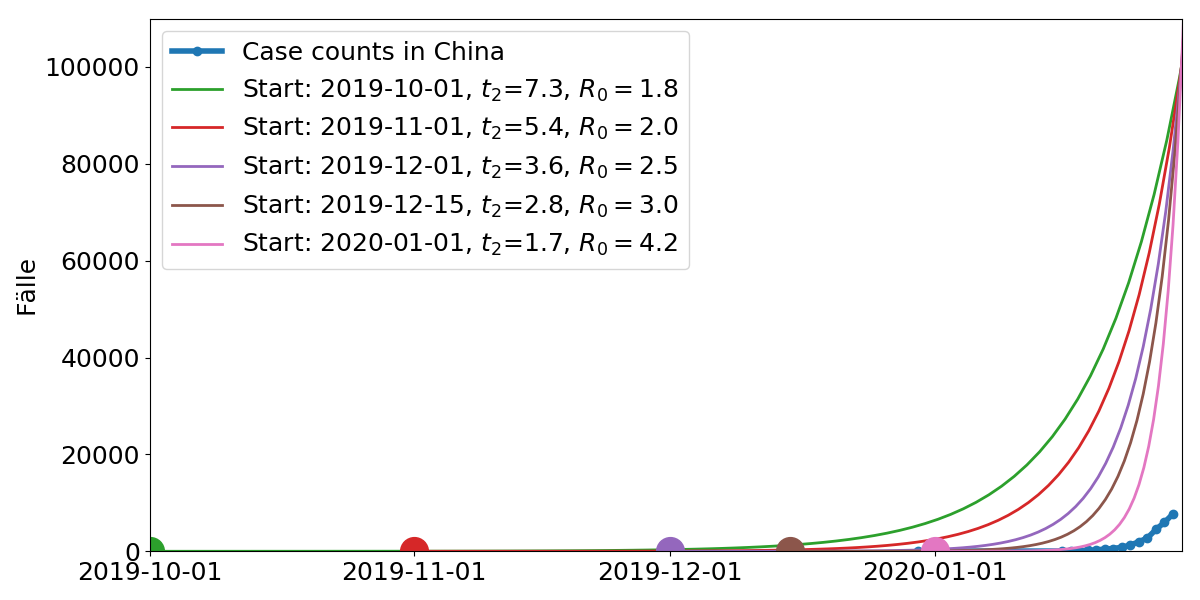
Model parameters: Start time, doubling time $t_2$. $R_0$ is the average number of subsequent cases

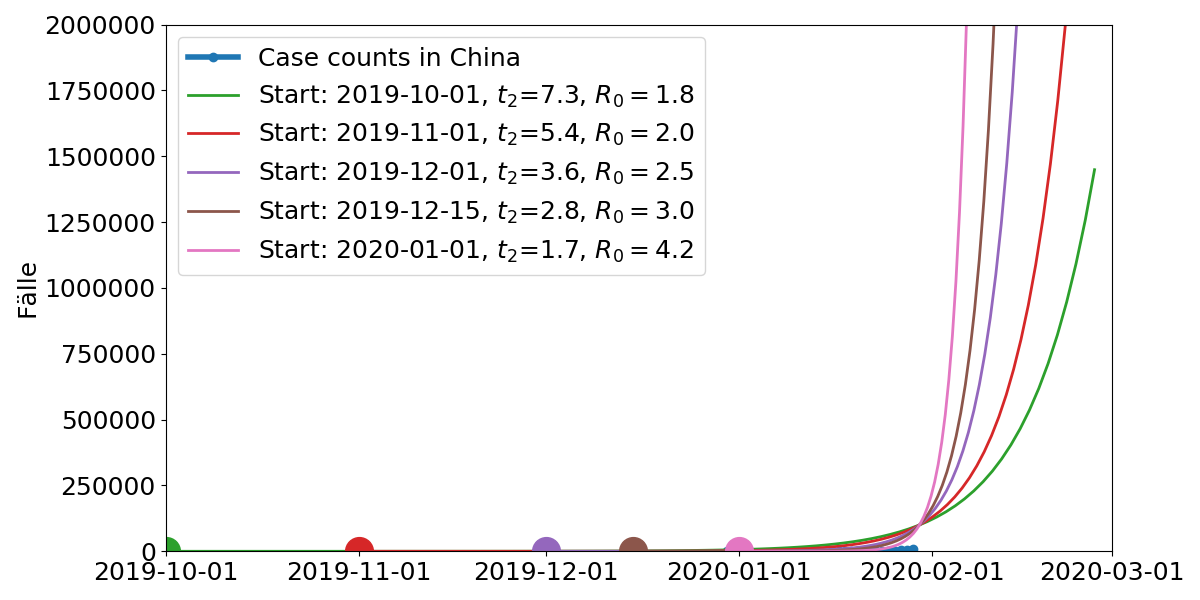
Model parameters: Start time, doubling time $t_2$. $R_0$ is the average number of subsequent cases

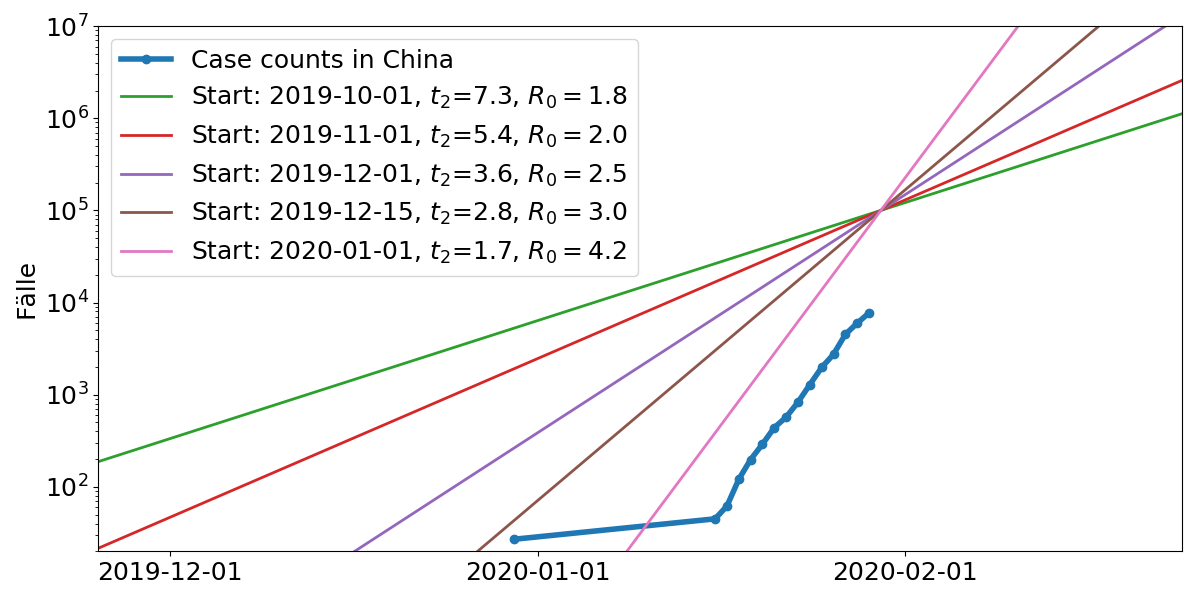
What are we dealing with here?
What can science contribute?

- Coronavirus: Diverse family of viruses
- Some variants lead to slight colds
- Others cause serious diseases (SARS, MERS)
- Many varieties circulate in bats.
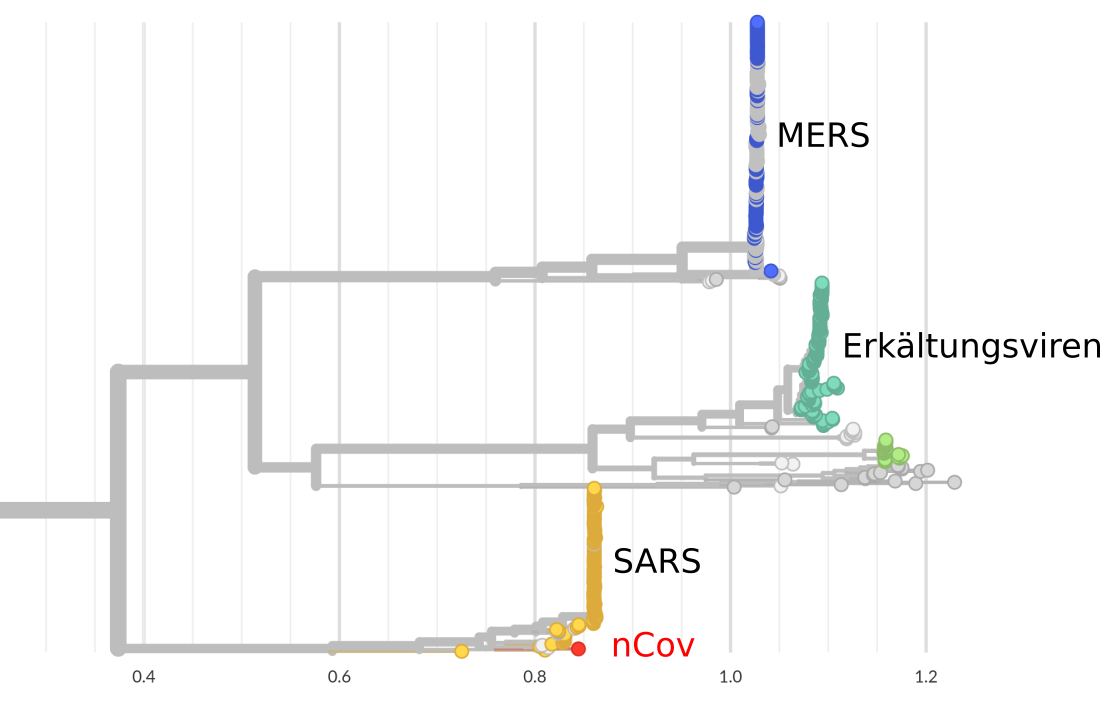
Virus genomes are constantly changing

Virus 1
CCATGAAGACTATCATTGCTTT...
Virus 2
CCATGAAGACTATTATTGCTTT...
Virus 3
CCATGAAGACTATCATTGCTTT...
CCATGAAGACTATCATTGCTTT...
Virus 2
CCATGAAGACTATTATTGCTTT...
Virus 3
CCATGAAGACTATCATTGCTTT...
All viruses mutate -- most mutations do not change the properties of the viruses!
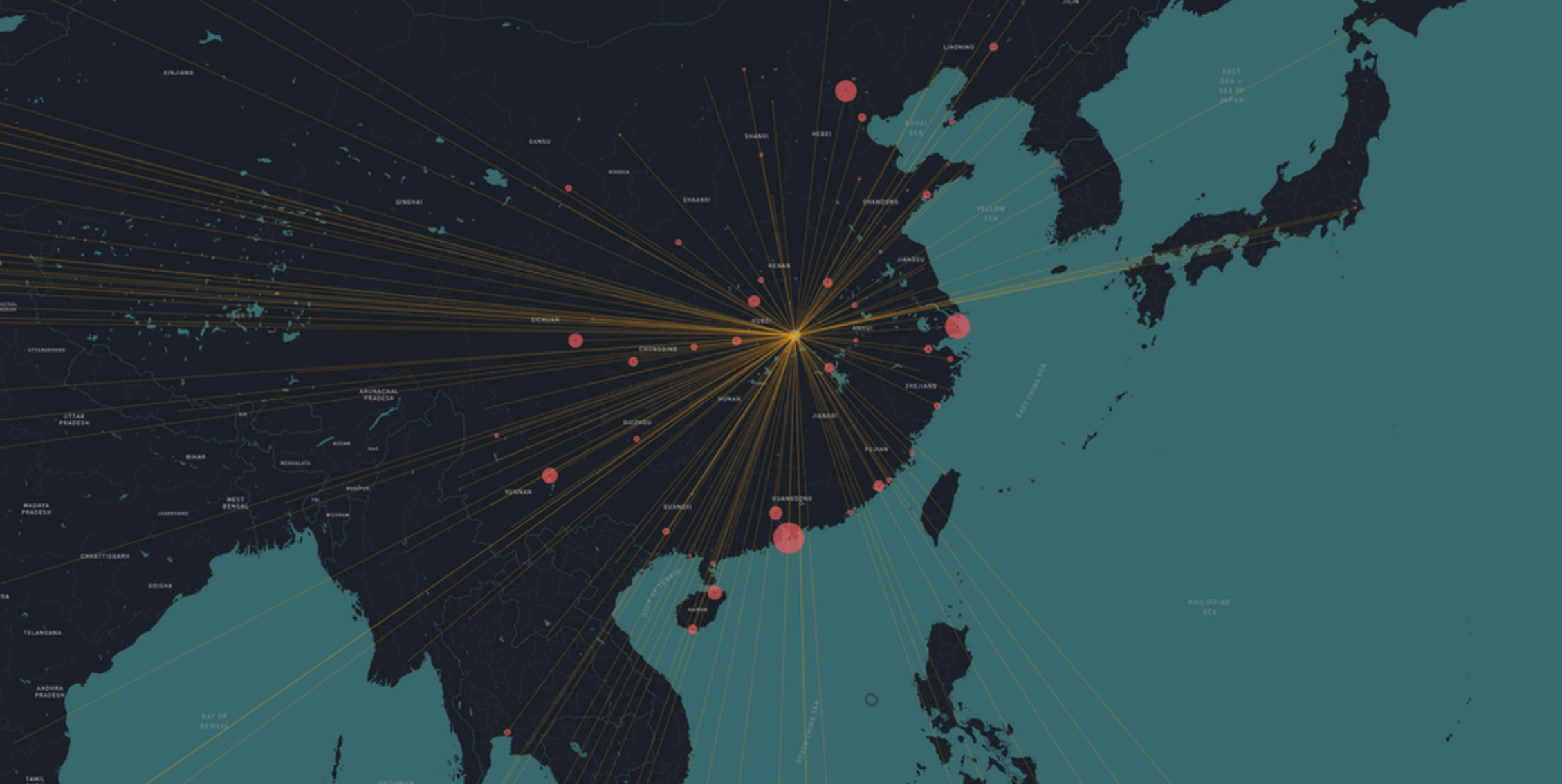
Mutations in the genome sequences document the spread





About 1-3 mutations per month
nextstrain.org
Joint project with the laboratory of Trevor Bedford (Seattle)
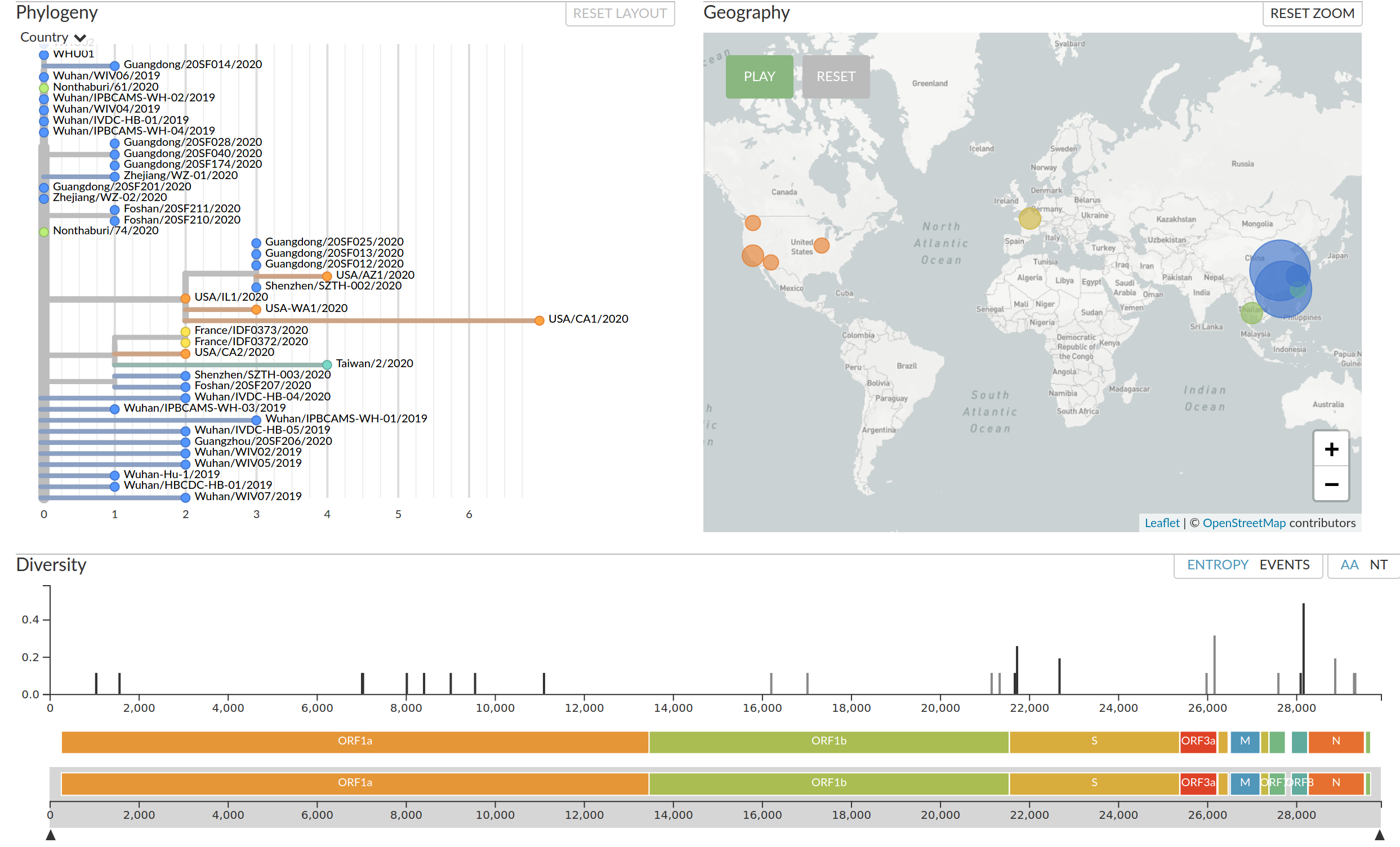
Epidemiological Key Points

-
Start of the outbreak: early December
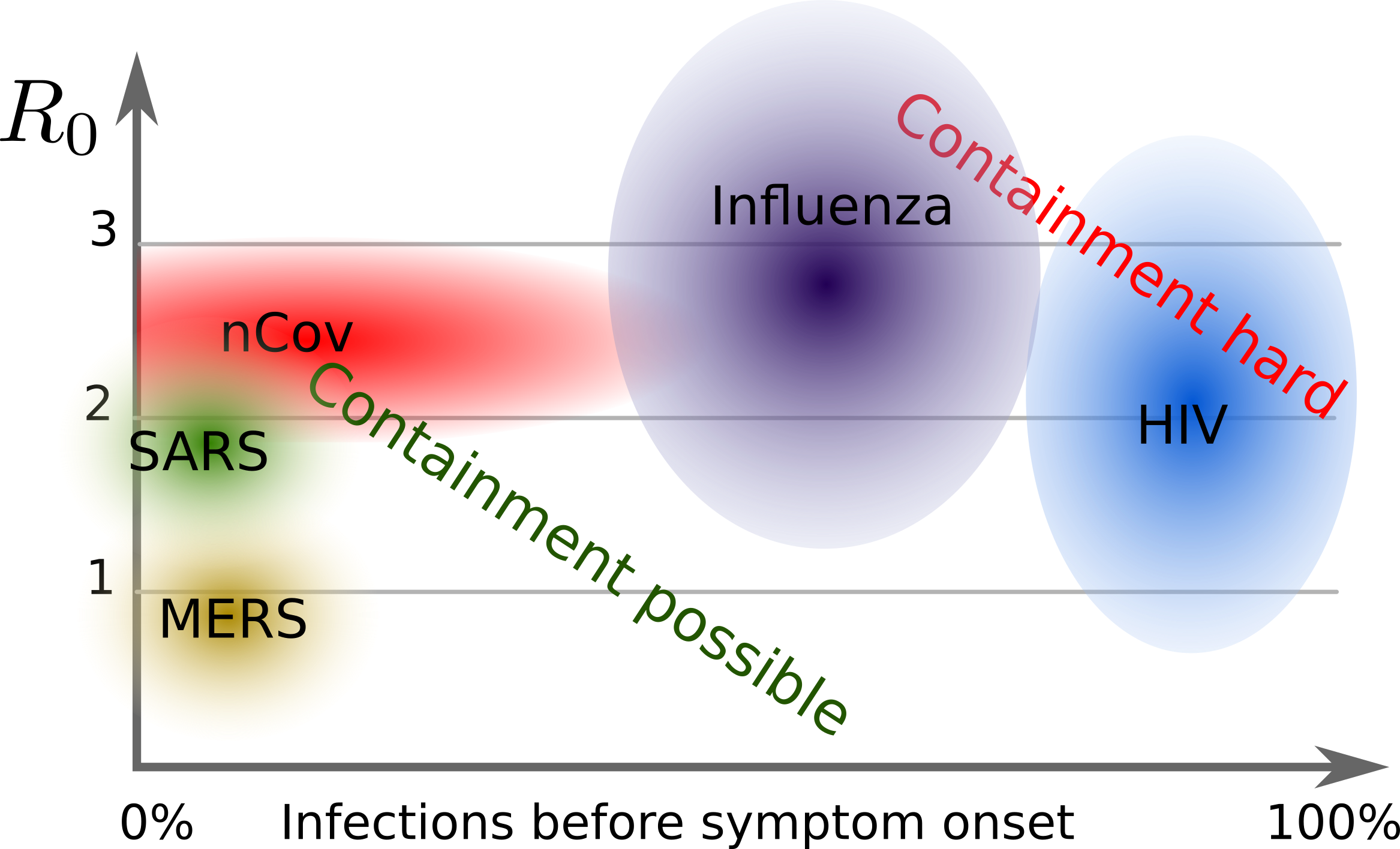
Growth rate: $R_0 = 2-3$ (Each infection leads to 2-3 subsequent infections)
→ Containment possible if 50-70% of all cases and contacts are quickly isolated.
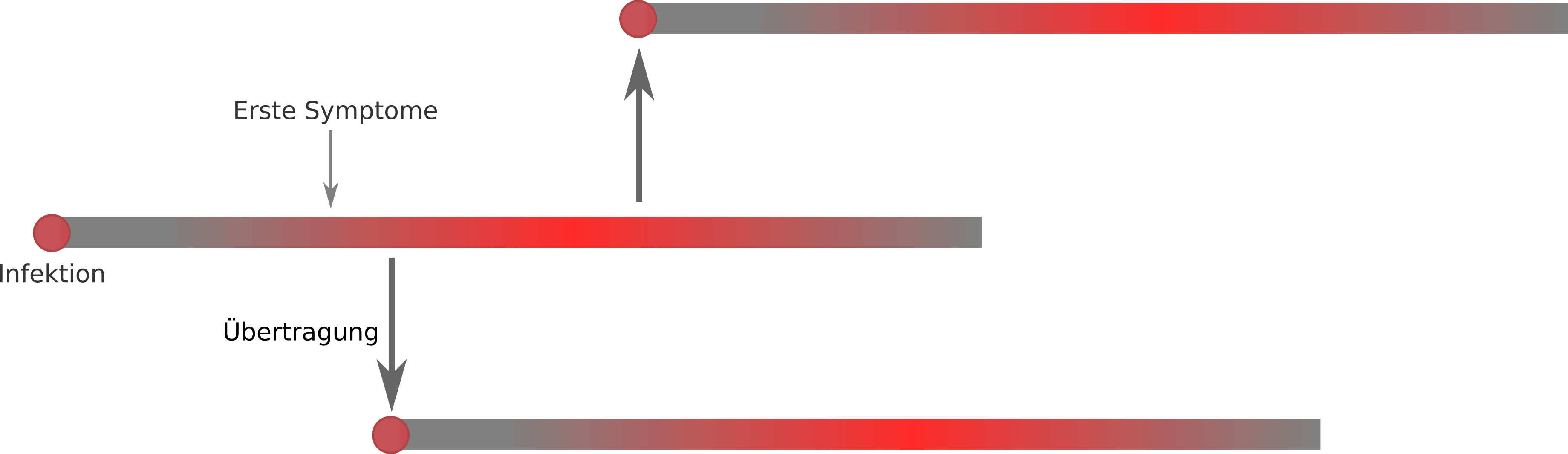
Extrapolation very difficult at the moment! → Comparison with historical outbreaks
2009-2010 -- Swine Flu (A/H1N1pdm)

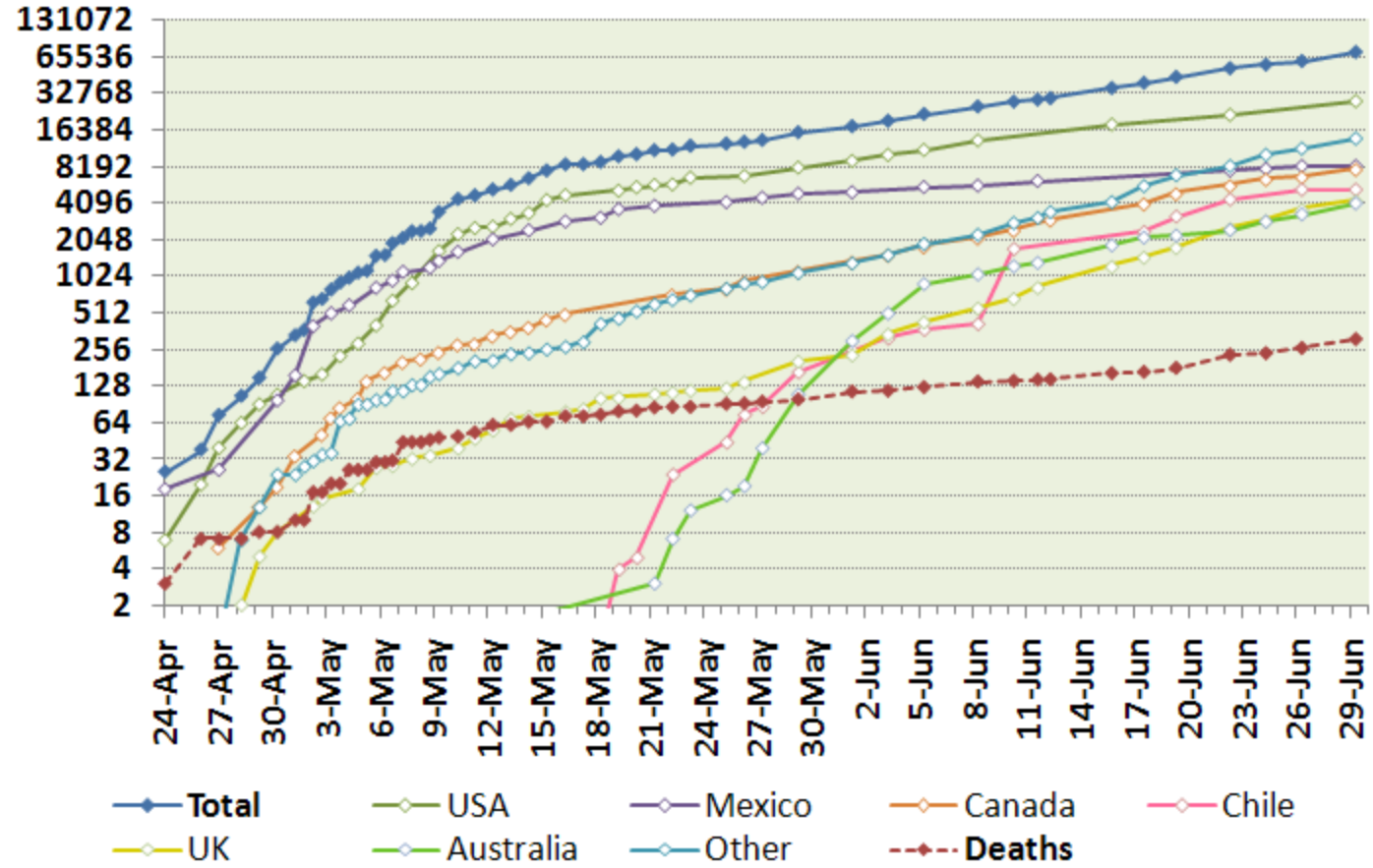
- Originated in Mexico in 2009
- Approximately 10% of the world population infected
- Mortality under 0.03%
Rapid spread -- fortunately few severe cases!
2003 -- SARS (Severe acute respiratory syndrome)



- November 2002 - July 2003:
about 8000 confirmed cases, 774 deaths - Related virus, probably originated in bats as well
- Epicenter in south China. Significant case numbers in Hong Kong, Taiwan, Singapore, Canada
- Mortality higher than nCov-2019
- Successful containment

Summary

- In China, the virus has been spreading unchecked so far.
- Origin multiplied end of November/beginning of December.
- Extrapolation of cases abroad indicates a total of 100,000 cases.
- Probably a large percentage of cases are mild.
- Imported cases can currently be well isolated and monitored.
- Containment would be more difficult if asymptomatic transmission is possible.
Acknowledgments



- Trevor Bedford
- Emma Hodcroft
- James Hadfield
- and others
- Medical staff, scientists, health authorities, and all others who care for the sick.
- WHO, GISAID, and other organizations for coordinations and data exchange.


