Real-time tracking of SARS-CoV-2 spread and evolution
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/202007_ISMB.html
Acknowledgments
Trevor Bedford and his lab -- terrific collaboration since 2014

especially James Hadfield, Emma Hodcroft, Ivan Aksamentov, Moira Zuber, and Tom Sibley
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID


 BBC
BBC
 Data summarized by Ian MacKay
Data summarized by Ian MacKay
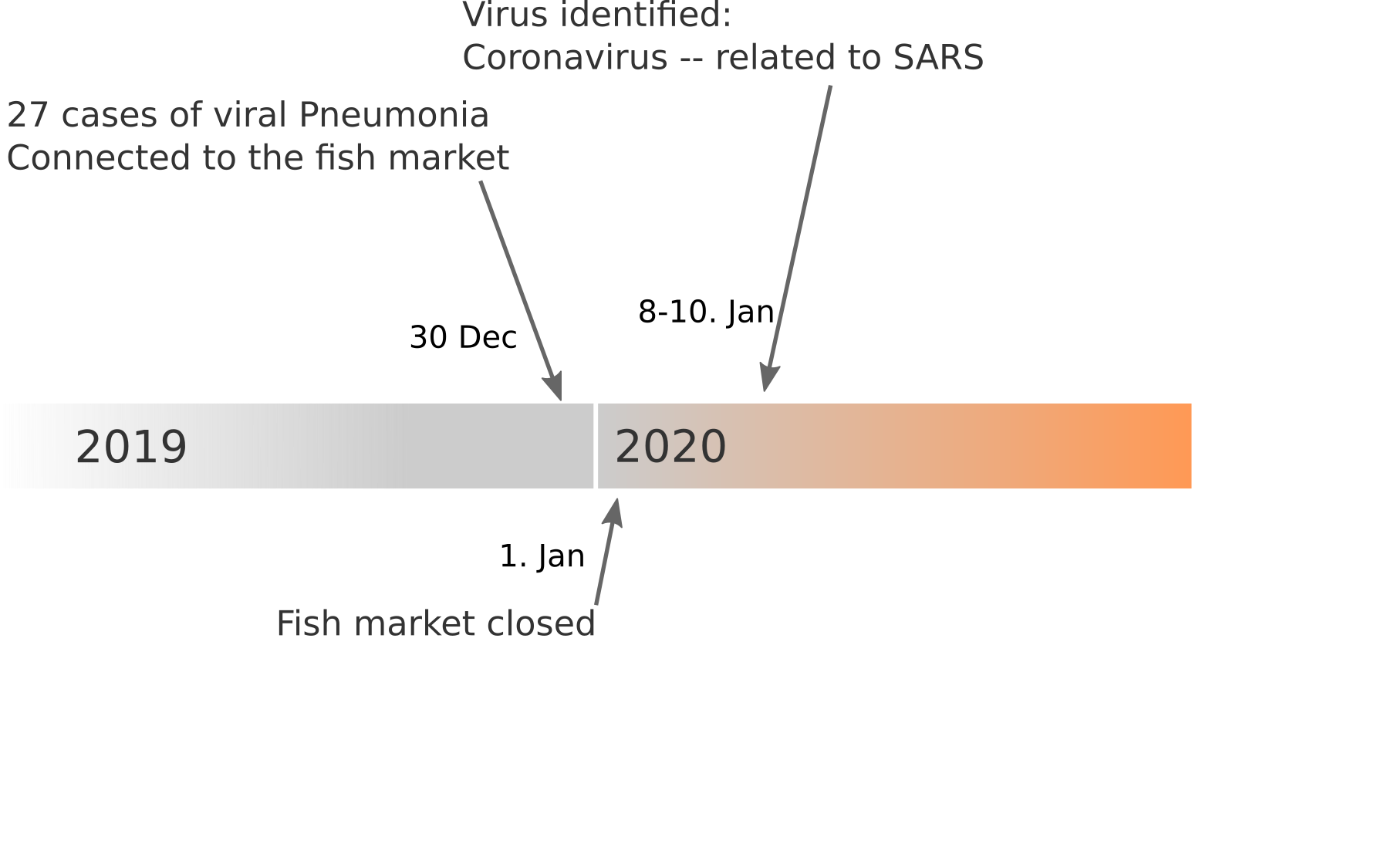 Data summarized by Ian MacKay
Data summarized by Ian MacKay
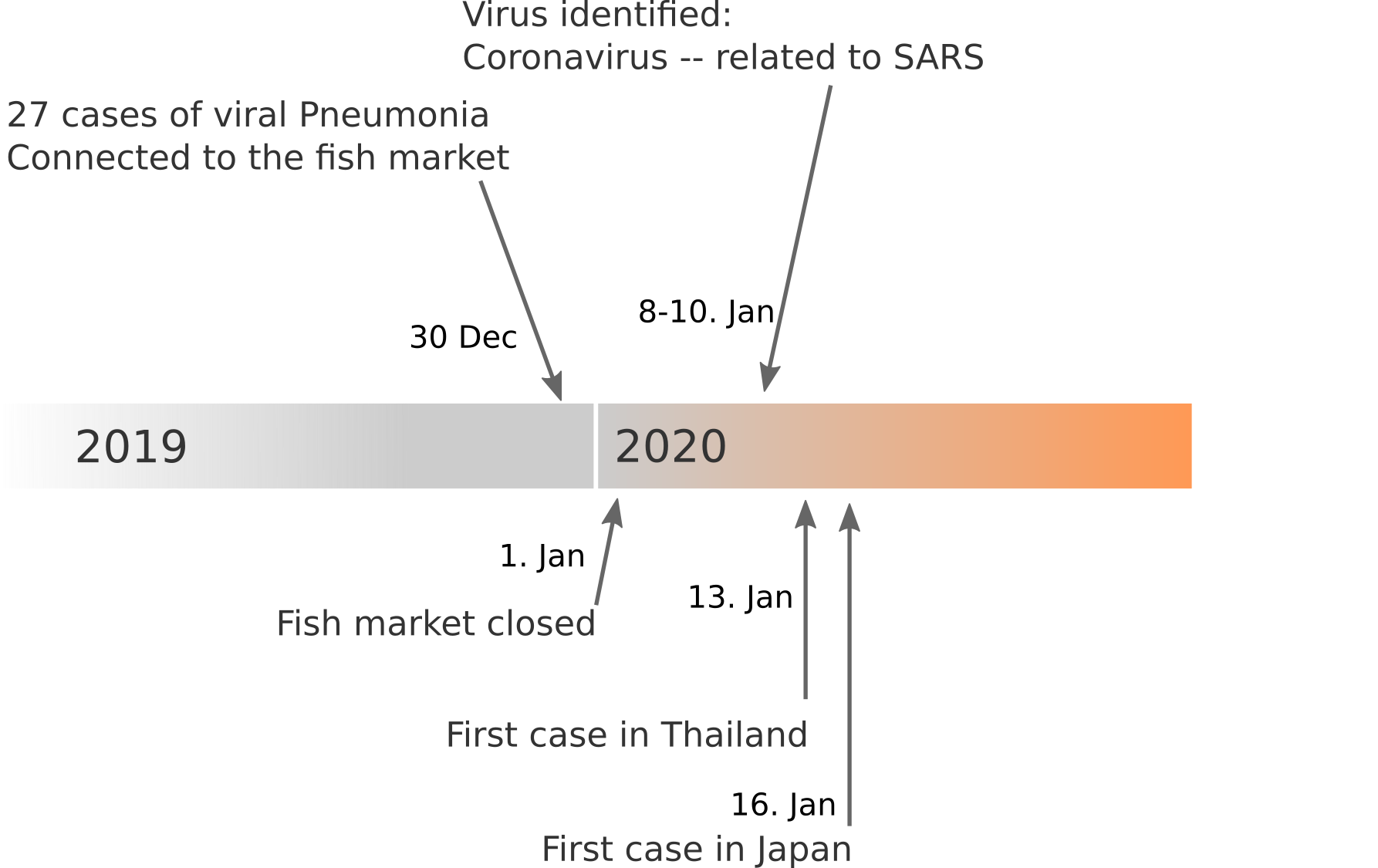 Data summarized by Ian MacKay
Data summarized by Ian MacKay
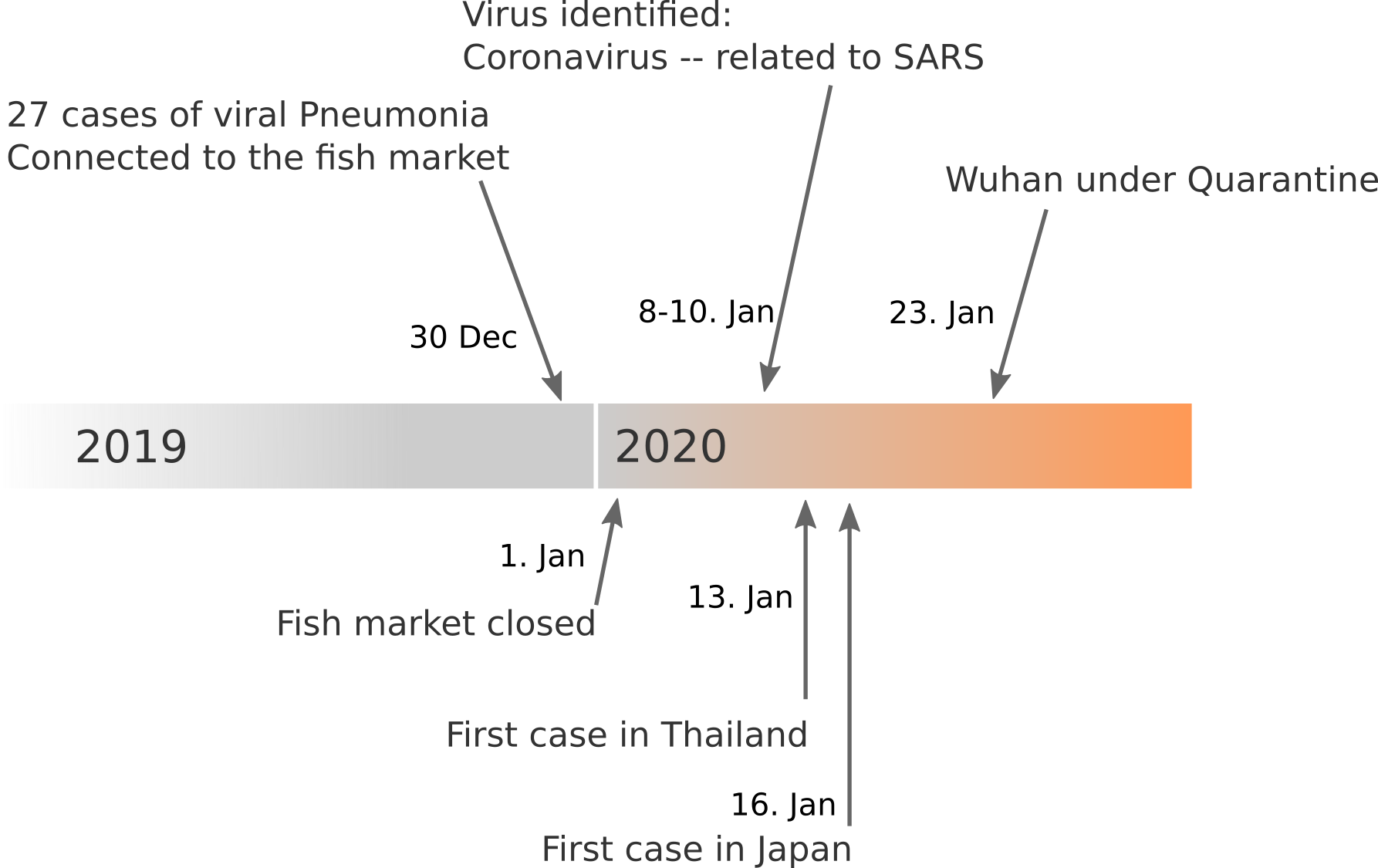 Data summarized by Ian MacKay
Data summarized by Ian MacKay
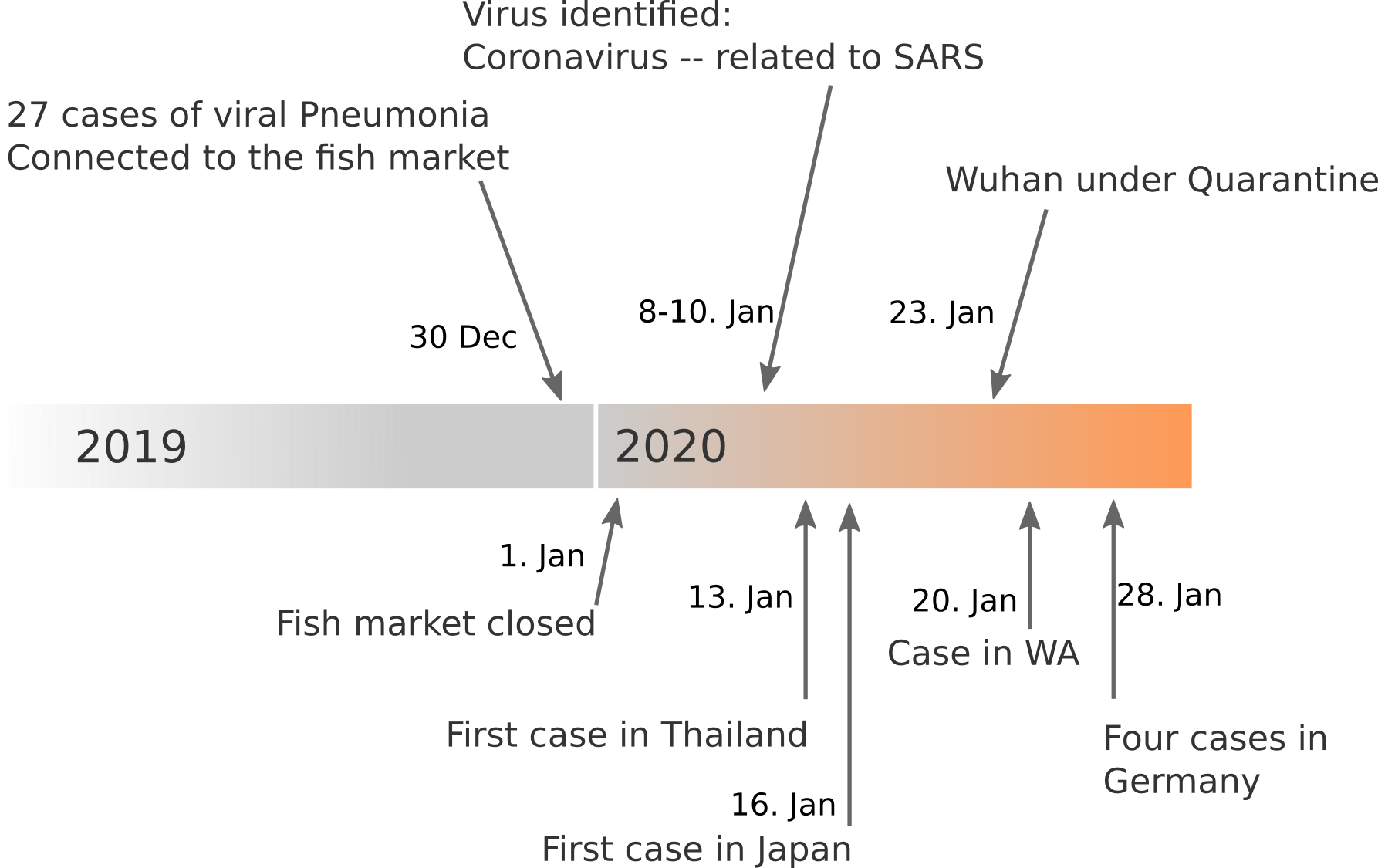 Data summarized by Ian MacKay
Data summarized by Ian MacKay
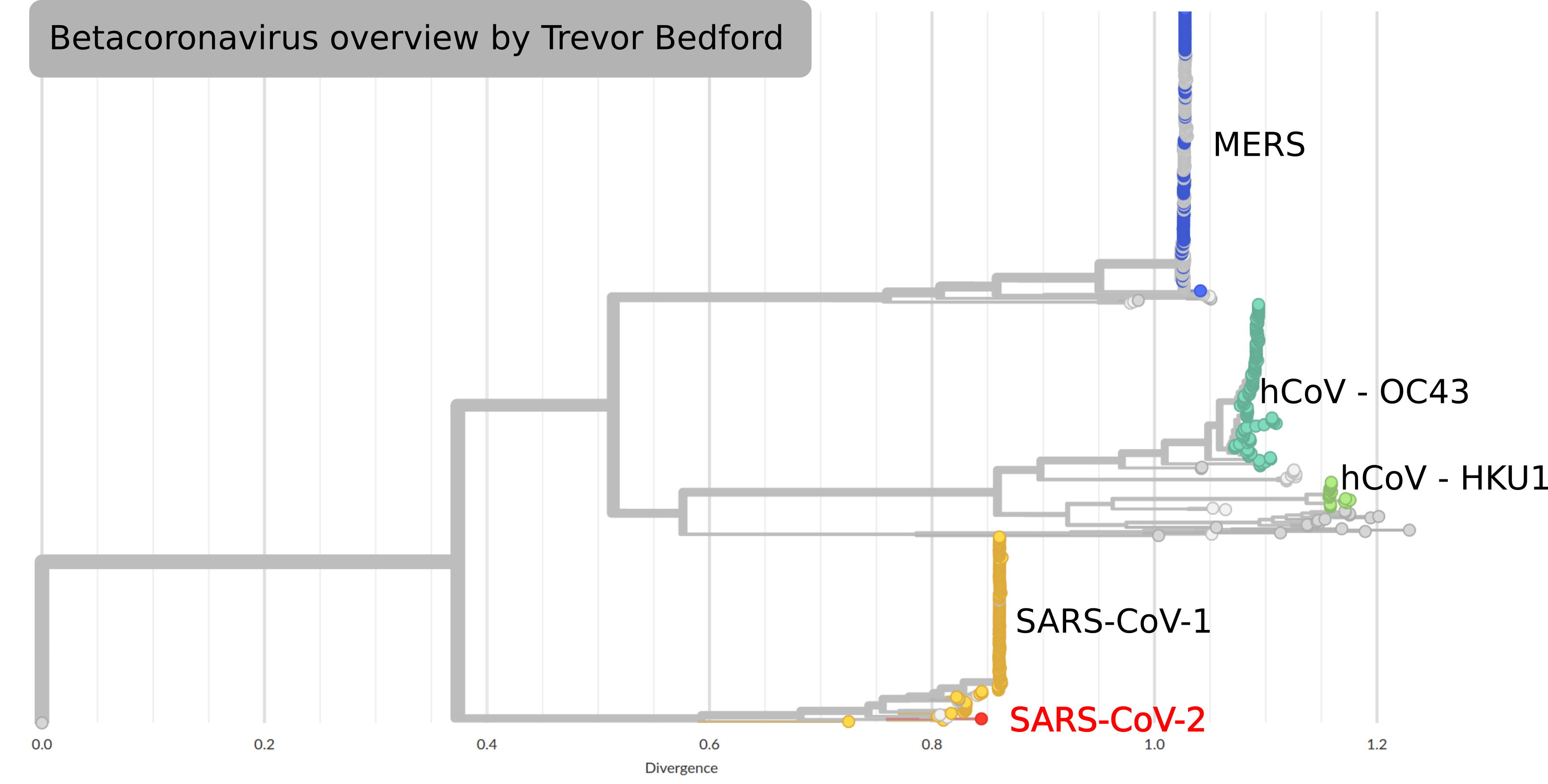 by Trevor Bedford
by Trevor Bedford
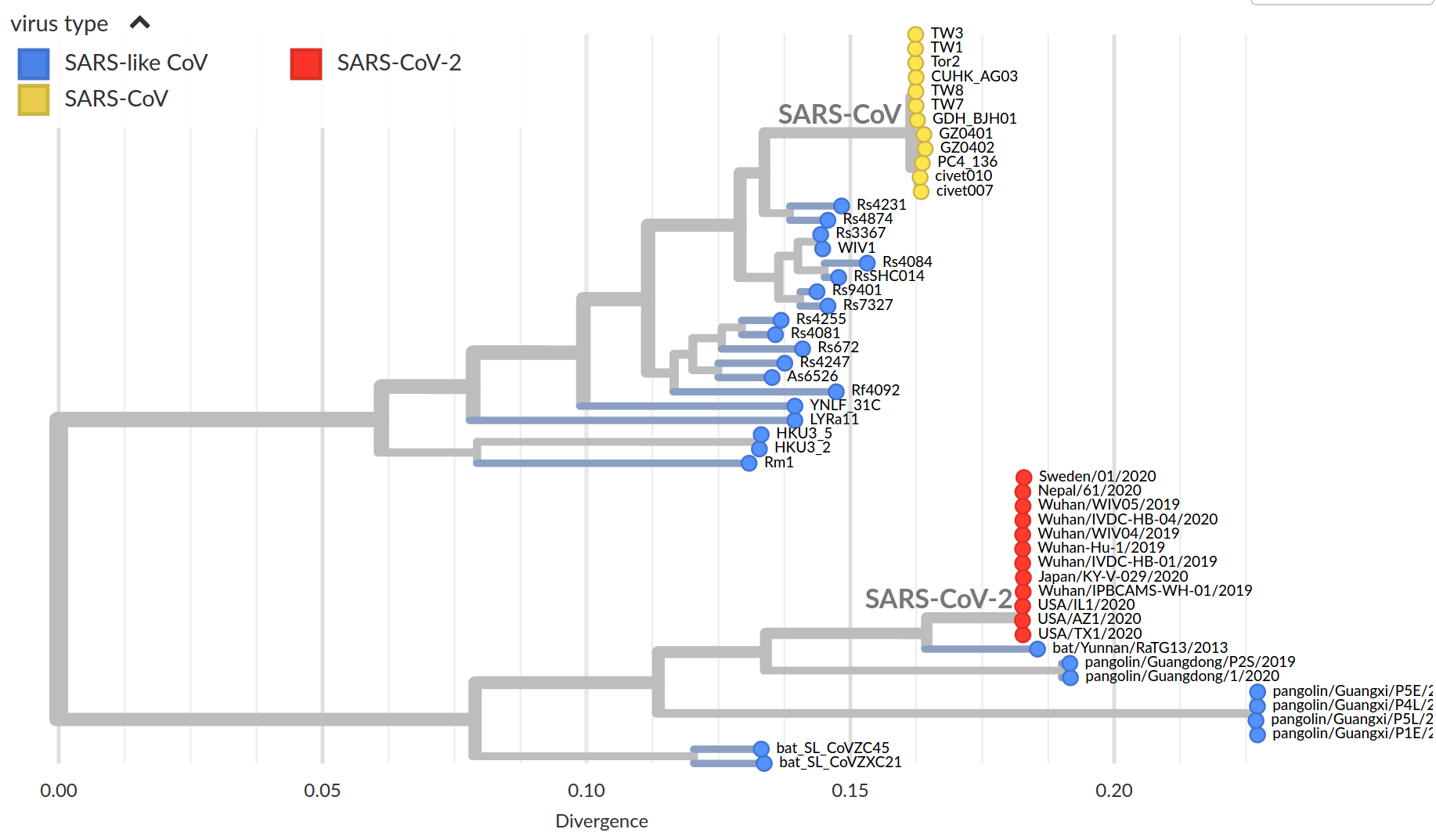 by Trevor Bedford
by Trevor Bedford
SARS-CoV-2 and its relatives
- SARS-CoV-2 is in the same family as SARS-CoV-1, MERS and
two seasonal coronavirus - The latter cause mild disease, the former severe
- Relatives are found in many different animals
- Closest known relative of SARS-CoV-2 is a virus isolated from bats
(RaTG13, approx 96% identical) - SARS-CoV-2 spreads more easily than SARS-CoV-1 or MERS;
pre-symptomatic transmission makes it hard to control
The SARS-CoV-2 genome
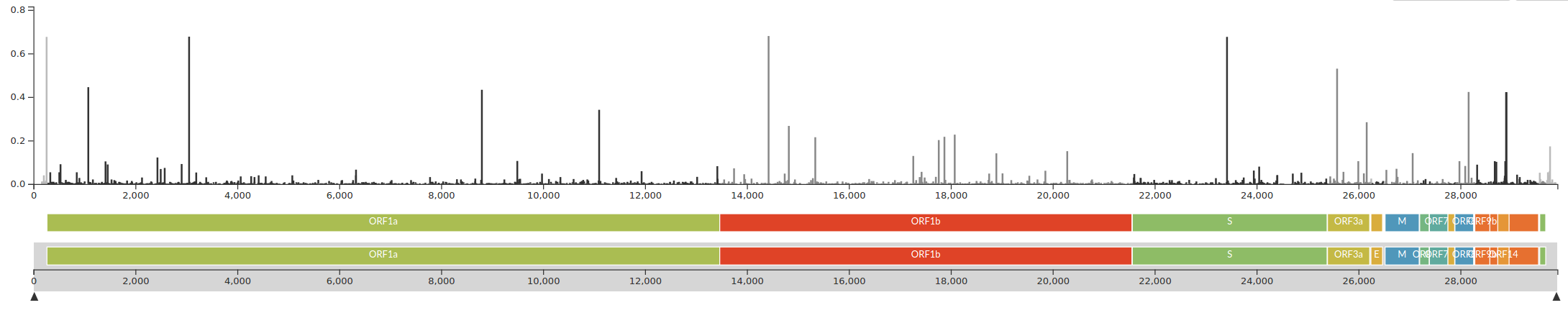
- 29k linear (+)ssRNA genome -- one of largest RNA virus genomes
- the first 2/3 code for the replication machinery
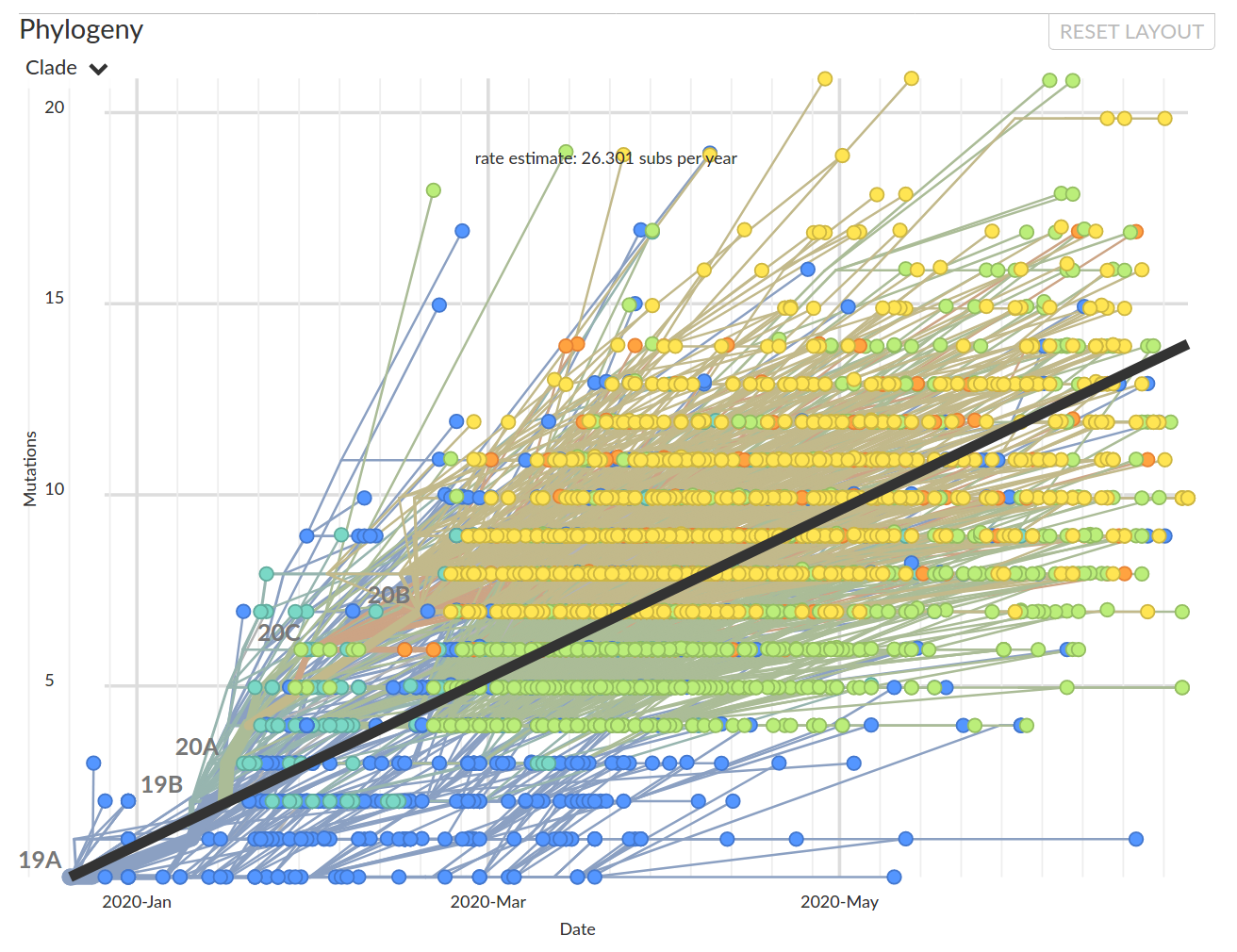
- While it spreads, it accumulates about 2 mutations per month
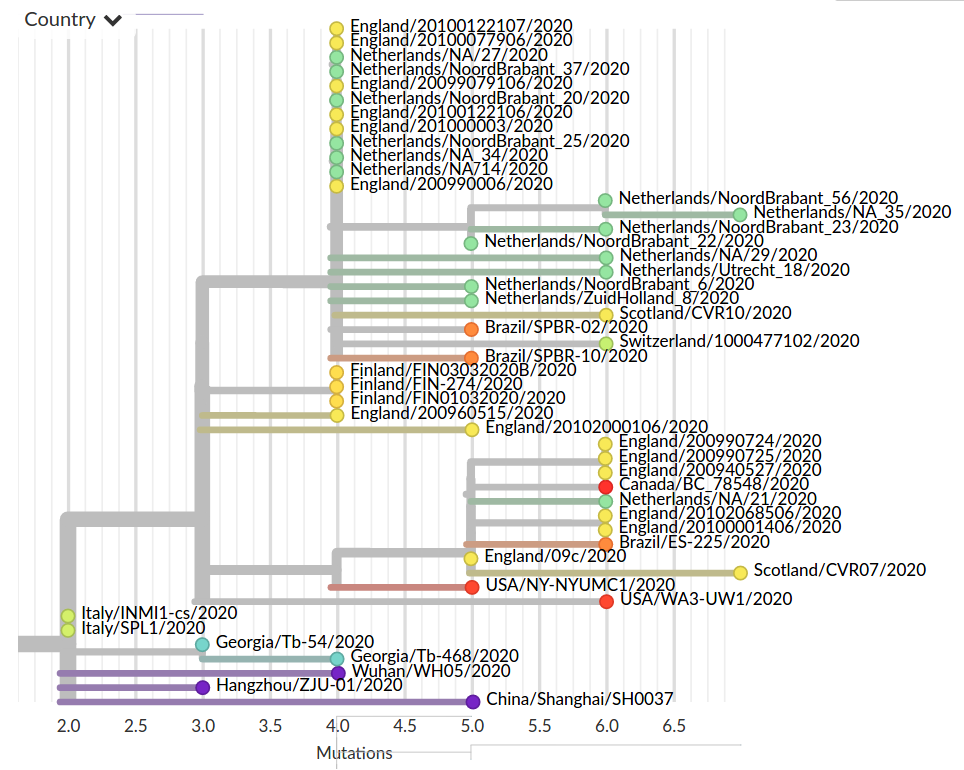
Tracking diversity and spread of SARS-CoV-2 in nextstrain
Available data on Jan 26
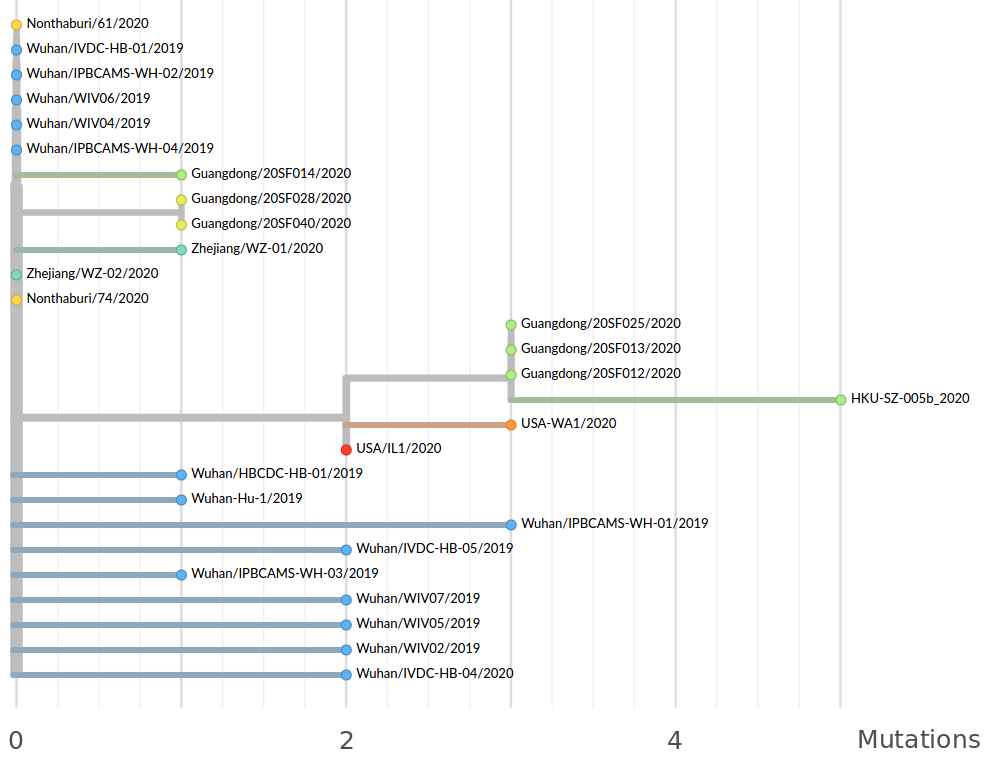
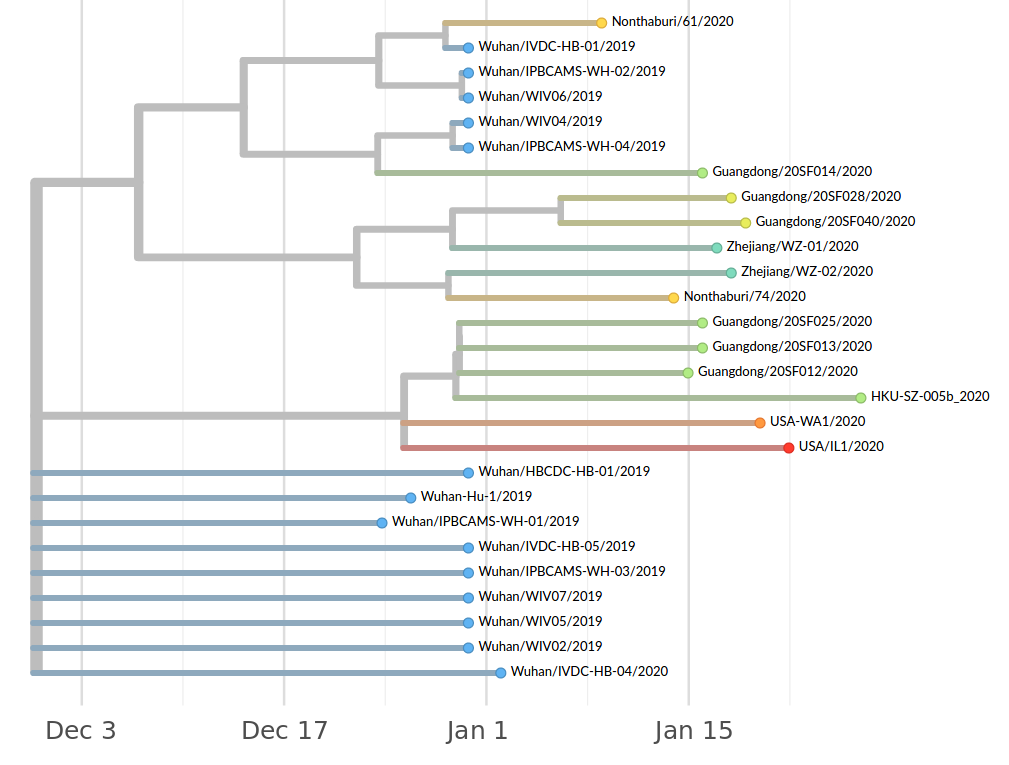
Early genomes differed by only a few mutations, suggesting very recent emergence
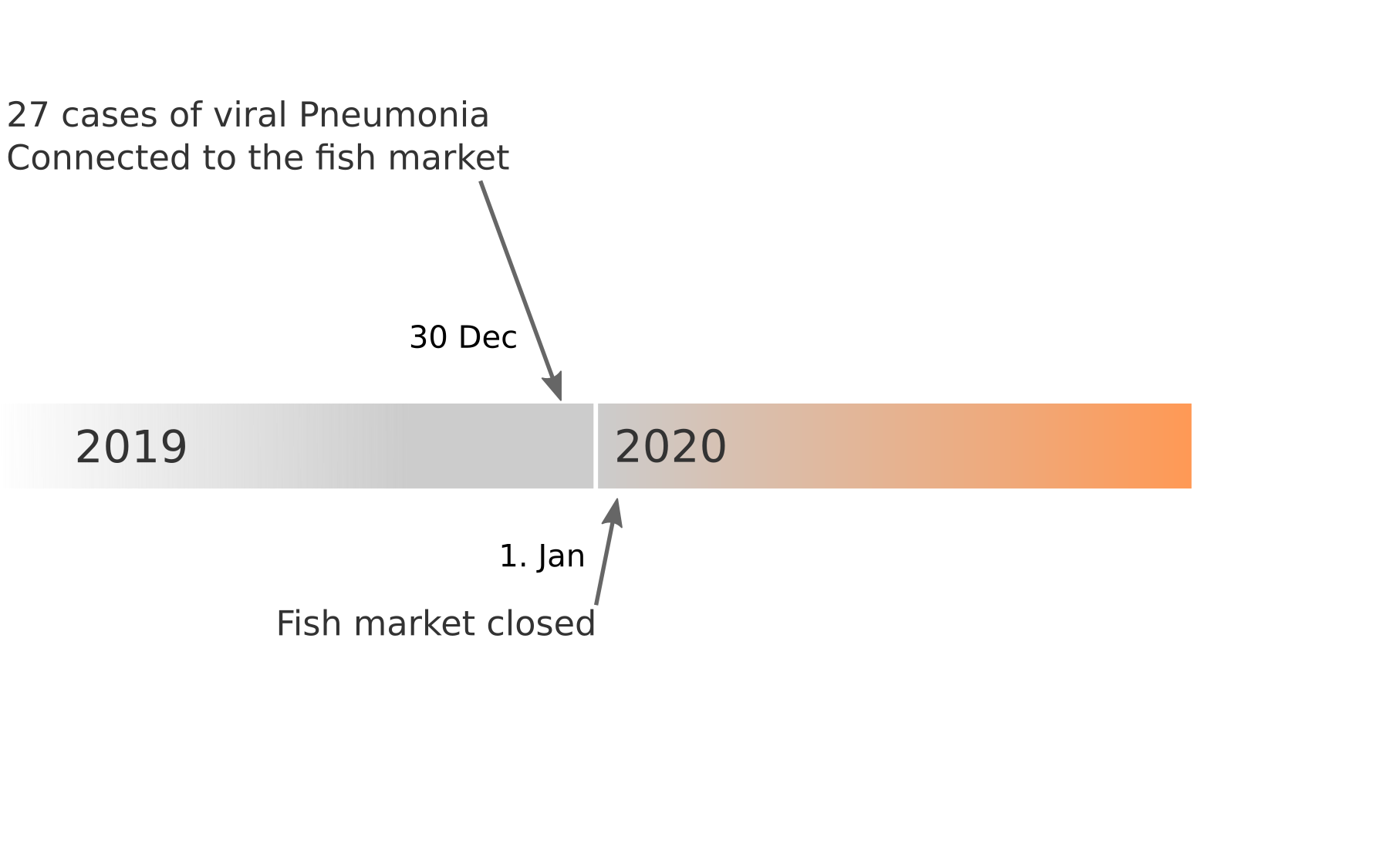
Early lessons from SARS-CoV-2 genomes (mid Jan)
- The outbreak originated from one source
→ not repeated zoonoses from a diverse reservoir. - The common ancestor of all samples was Nov - early Dec 2019.
- Family clusters showed up as identical genomes (expected)
- A second clade emerged that will continue to spread
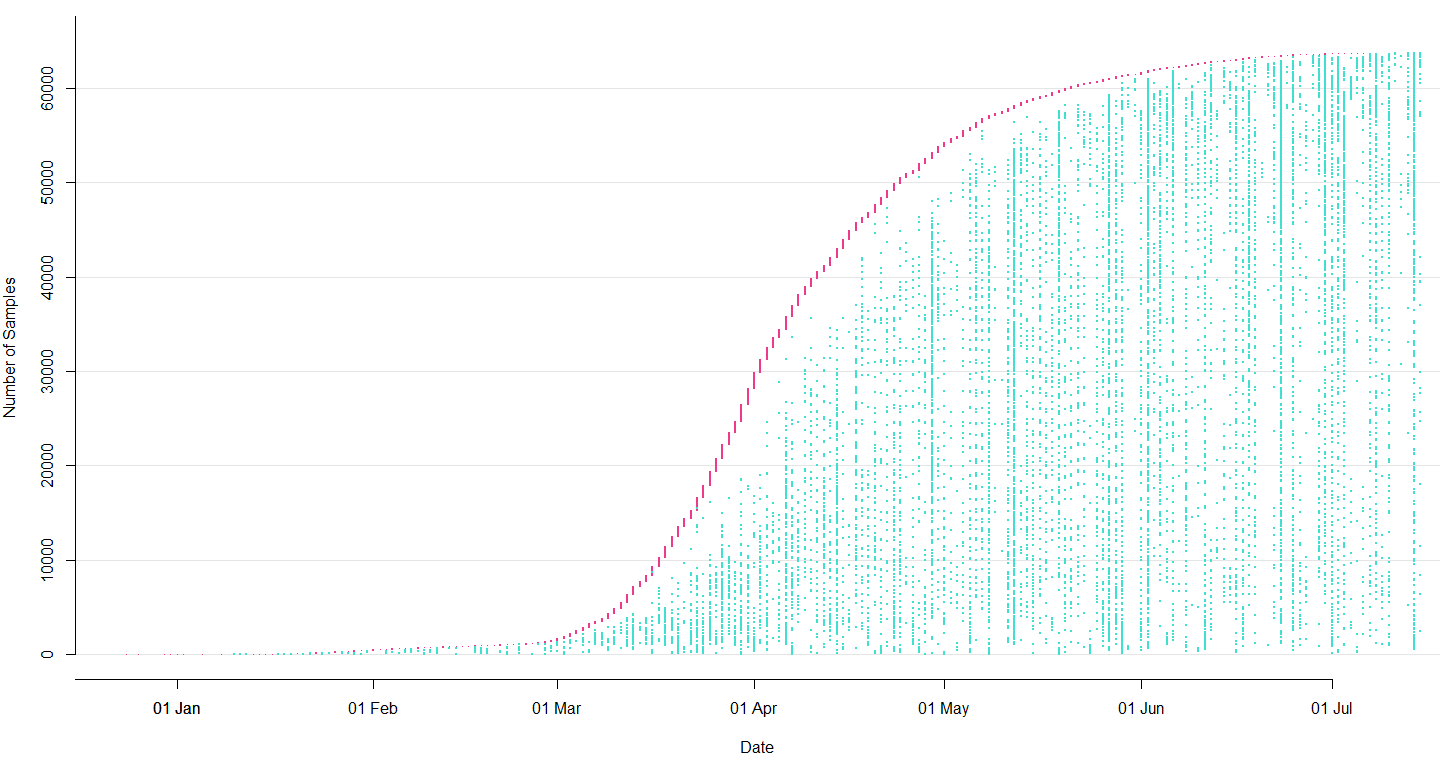
→ the closest to "real-time" we have experienced so far...
Figure by James Hadfield/Emma HodcroftSubset on July 14
As SARS-CoV-2 spreads, in accumulates on average 2 mutations per month
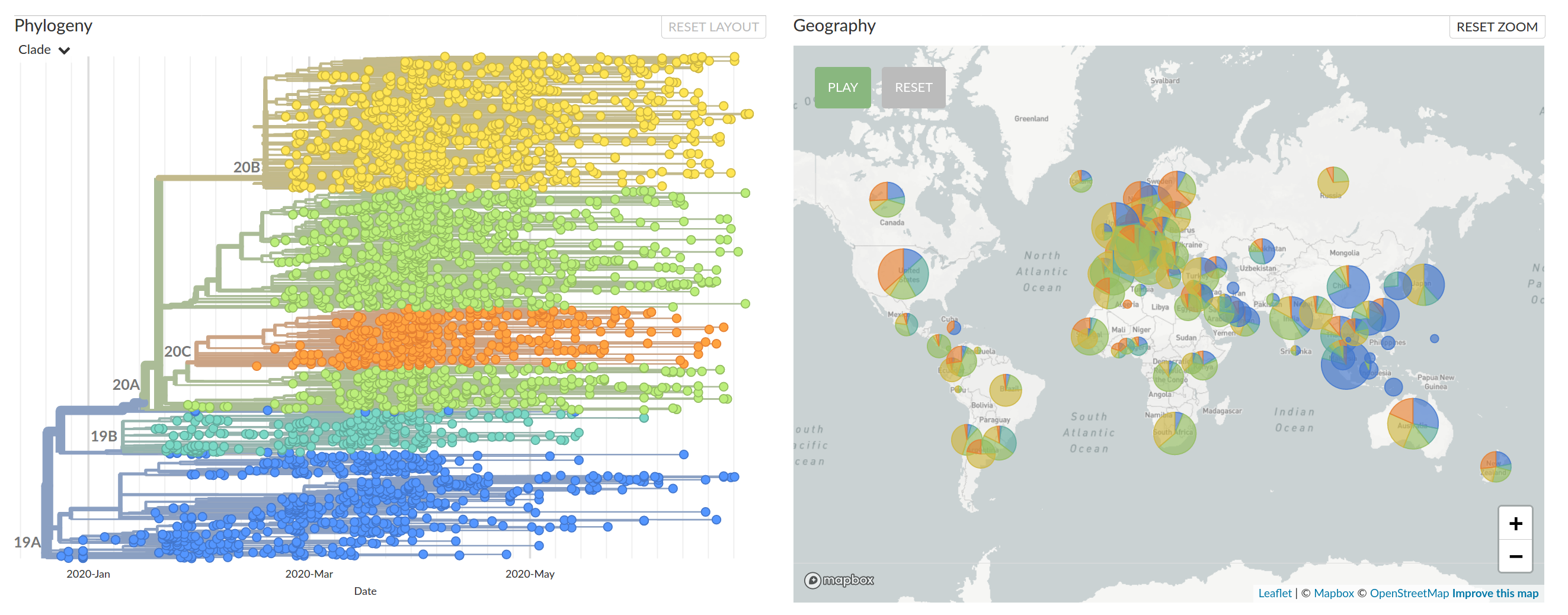
Tracking the spread of variants
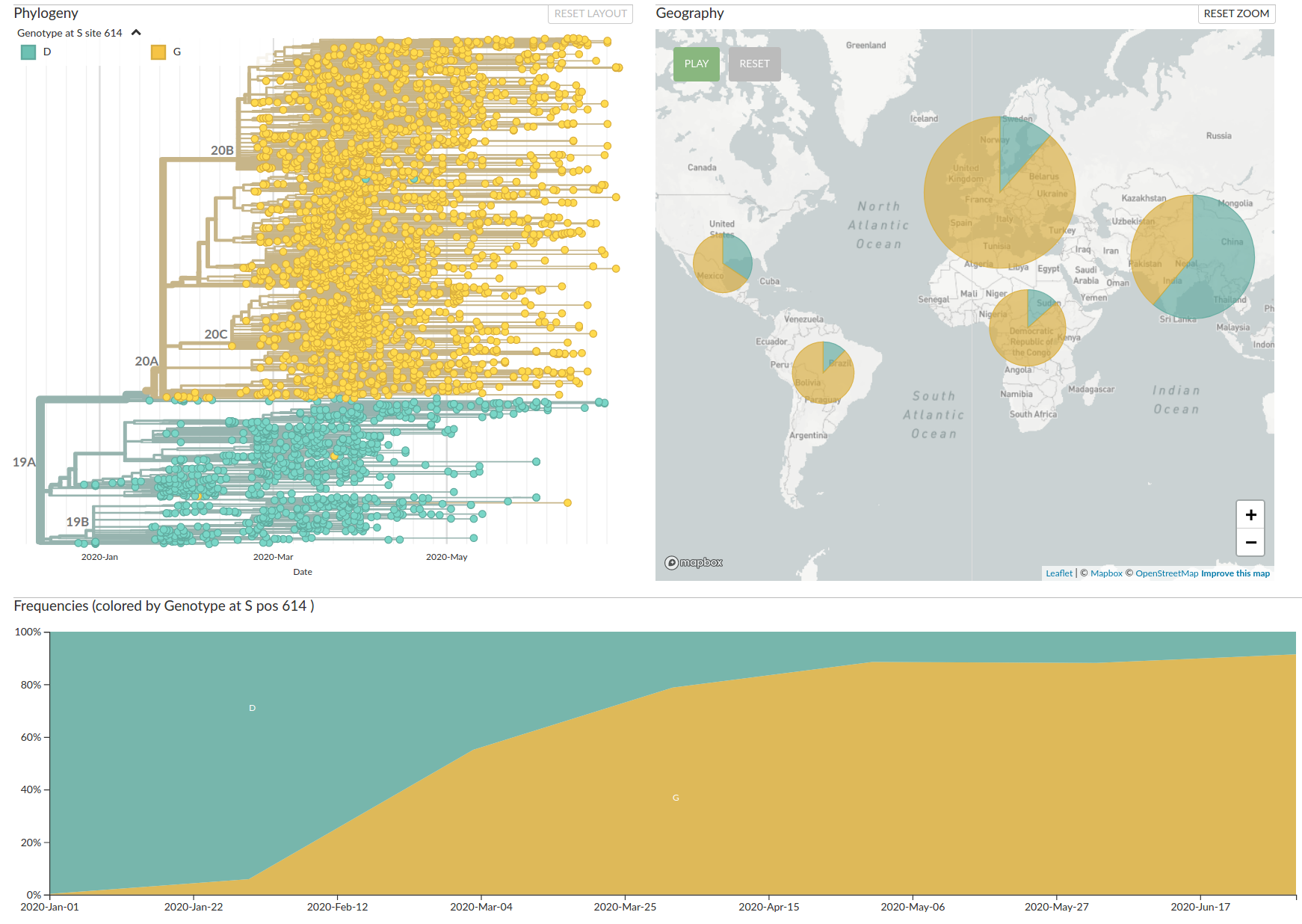
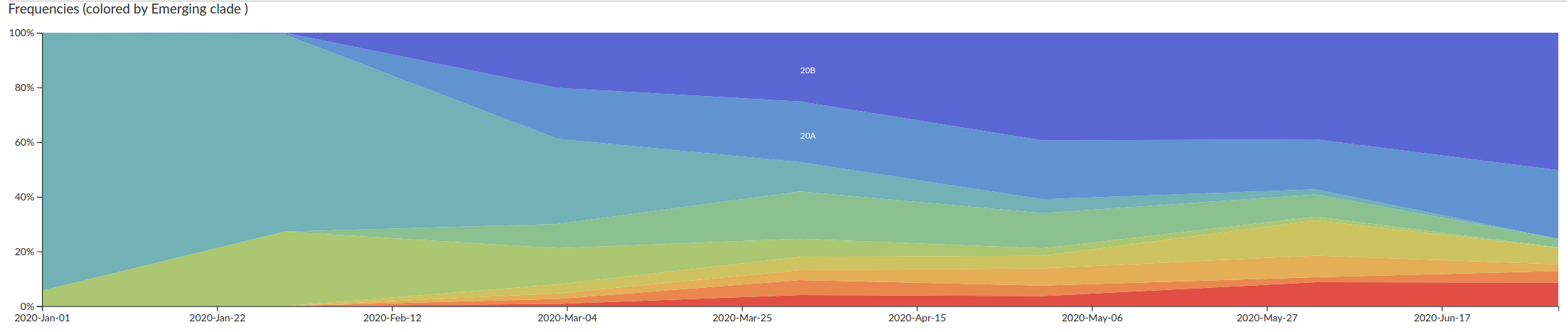
Tracking the spread of variants
- Clade 20A (and its daughters 20B and 20C) have essentially taken over
- With them, several mutations, including S:D614G have become prevalent.
- There are reports that S:D614G increases transmission
(equivocal in my view) - Overall no strong signal that the virus has changed in meaningful ways
- But mutations are very useful to reconstruct how the virus spreads
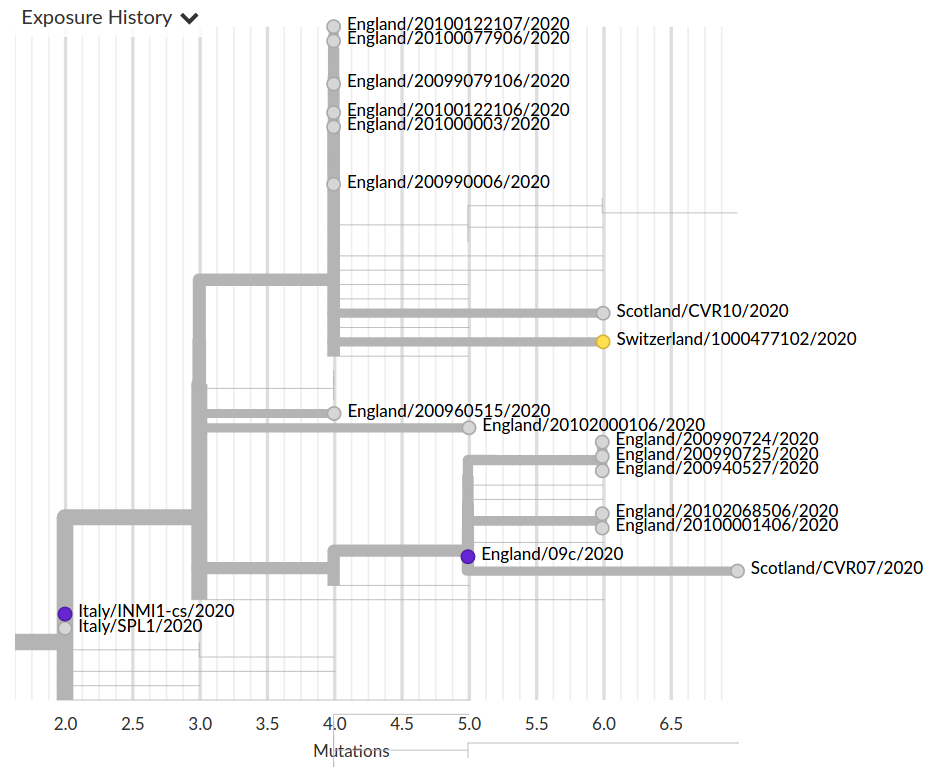
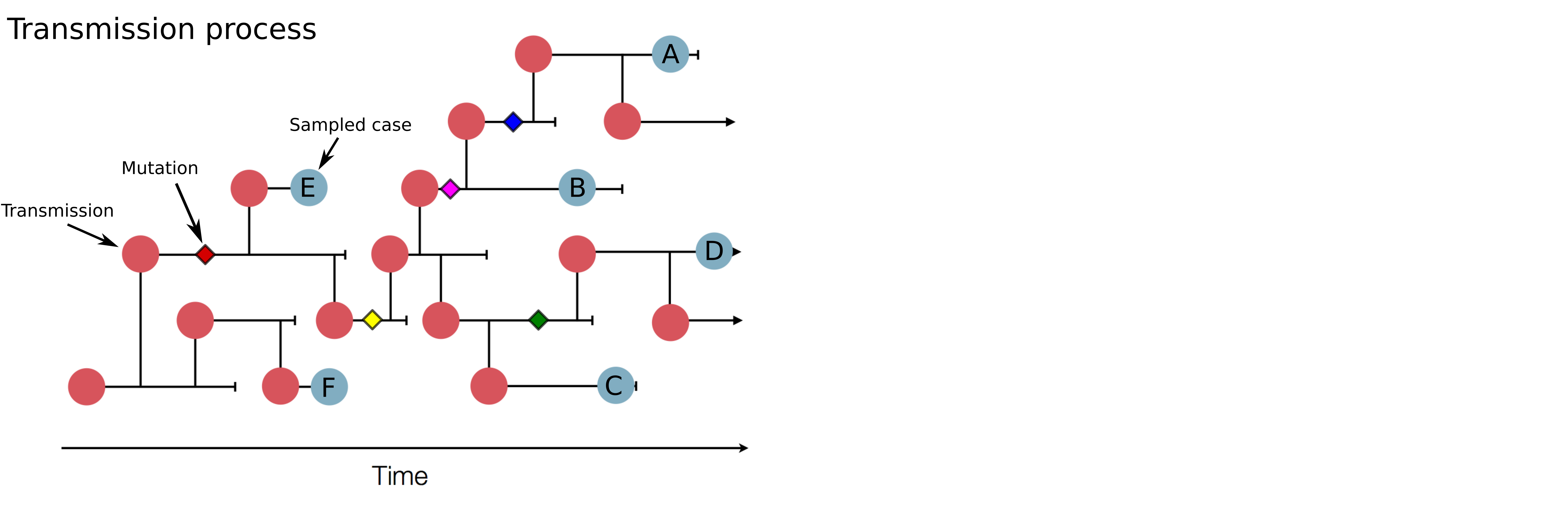
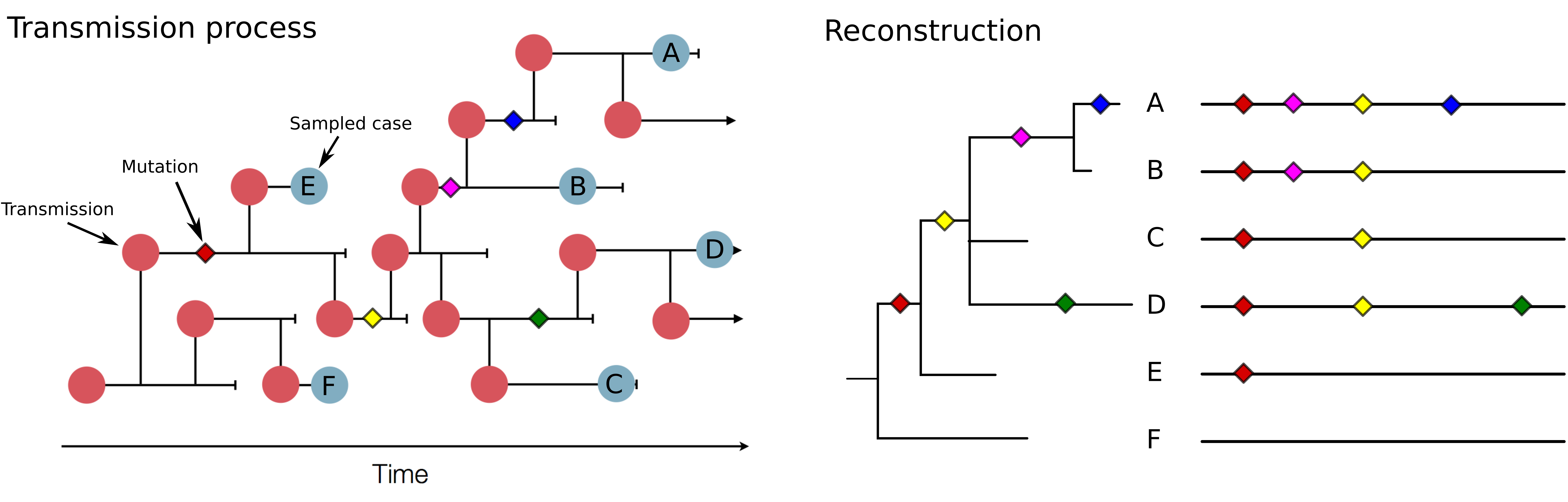
Genomic analysis as complement to contact tracing
Tracing the origins of samples from Iceland
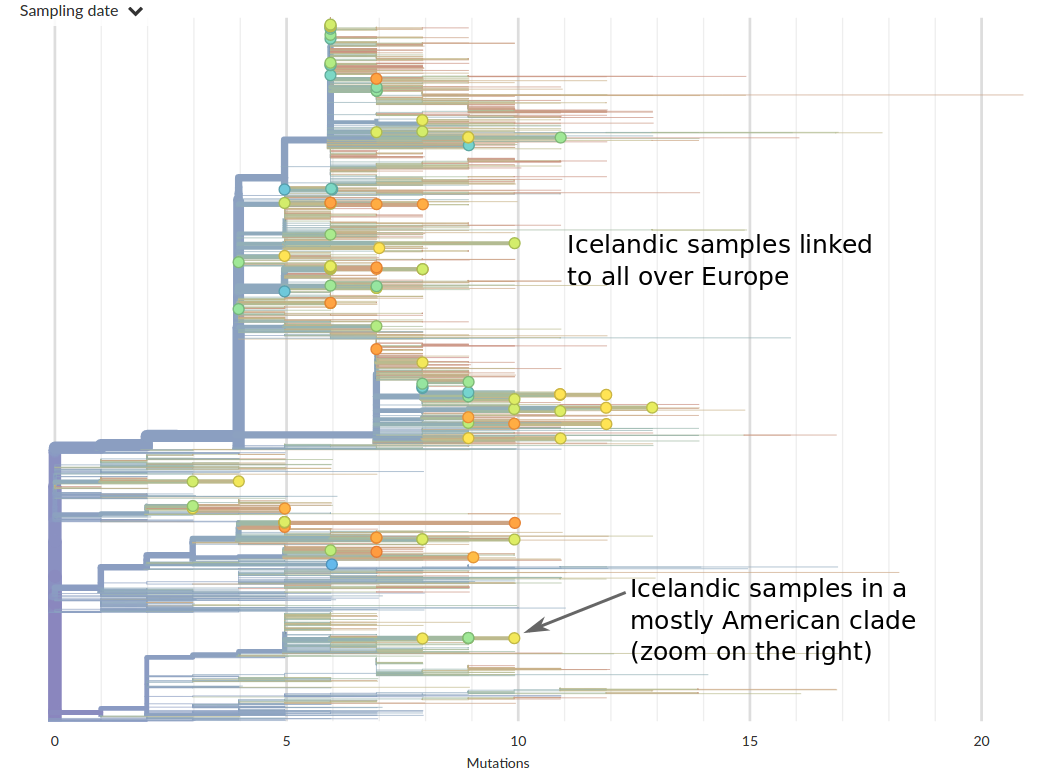
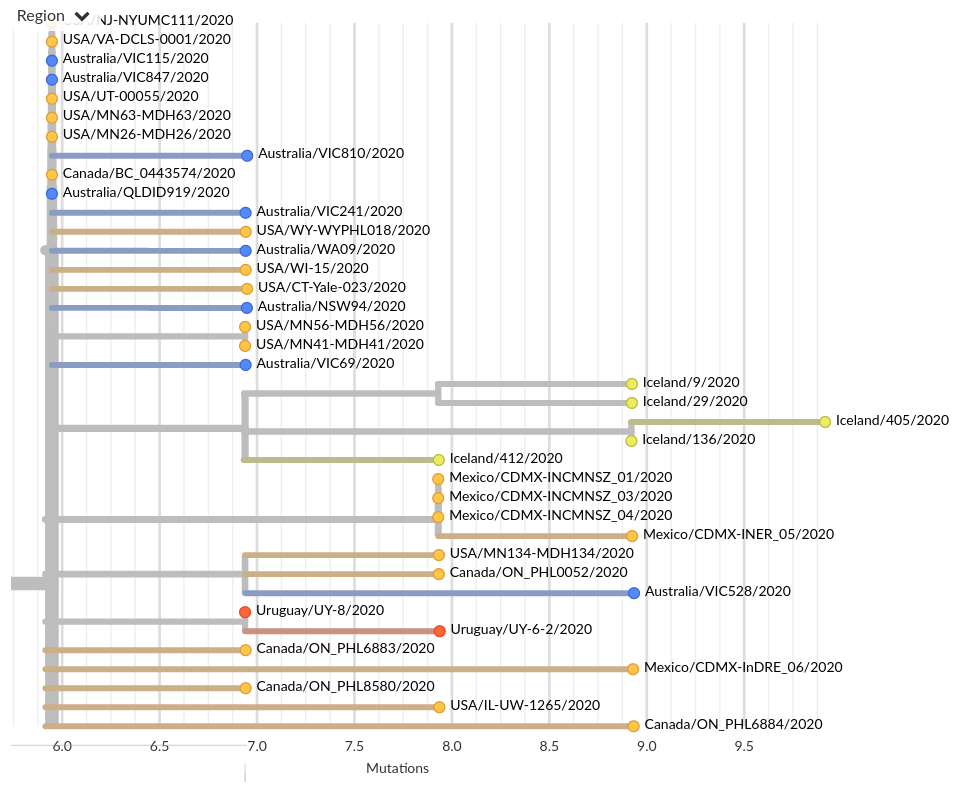
Prior to travel restrictions, SARS-CoV-2 is spread widely across the globe
Nextstrain was designed to track outbreaks in real-time ...
Initially:
Now:
- comprehensive global analysis kept up-to-date by us
- data curation done by GISAID
Now:
- Focus on local analyses by Public Health Departments
- Necessity to subsample sequences
- Often little experience with command line tools
- Large data sets → computational challenges
- Challenging to interpret
SARS-CoV-2 phylodynamic analysis with nextstrain
- input: metadata (csv table) + sequences
- composable snakemake pipeline
- filtering (select relevant genomes)
- alignment
- tree building
- infer time scaled phylogenies
- ancestral state reconstruction and phylogeography
- clade classification
- export to visualization
- runs in 1-3h for 4k genomes
Visualization with nextstrain
- run locally (localhost)
- deployed on your own servers
- shared via github hosting
- shared via nextstrain group (aws S3)
- sensitive metadata can be dropped onto the tree
- browser based tools to analyze your own sequences
What's next?
- Social distancing is very effective
→ we can suppress the outbreak if we want! - Many Asian and European countries have successfully suppressed the virus while re-opening
- What role does seasonality have?
(indoor vs outdoor activities)
Seasonal incidence of influenza viruses
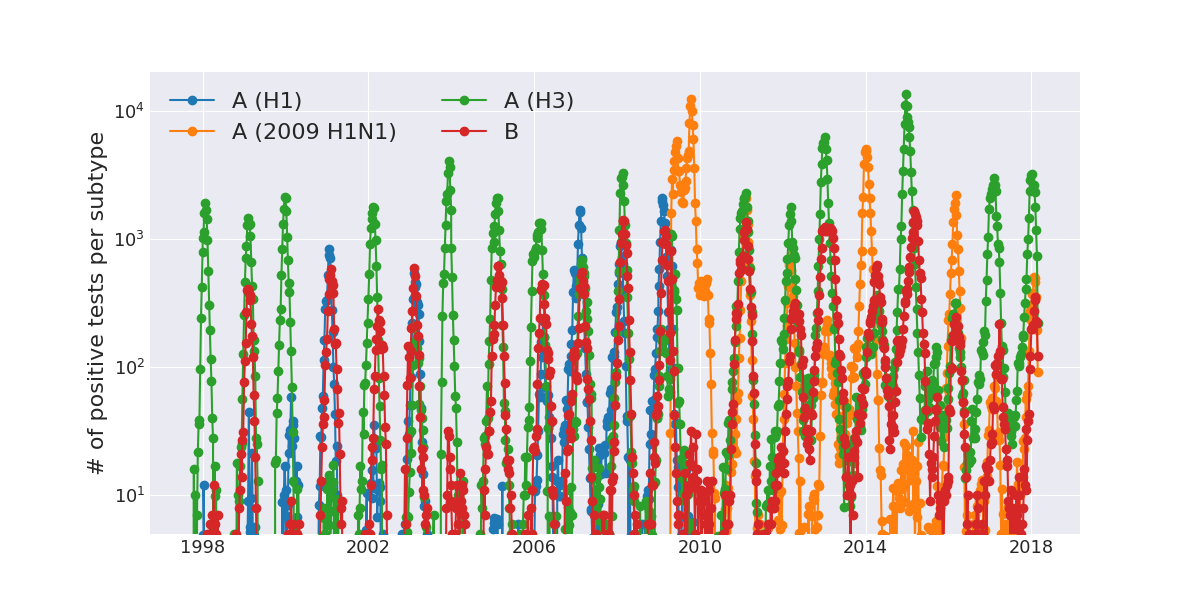
2009 pandemic influenza -- UK
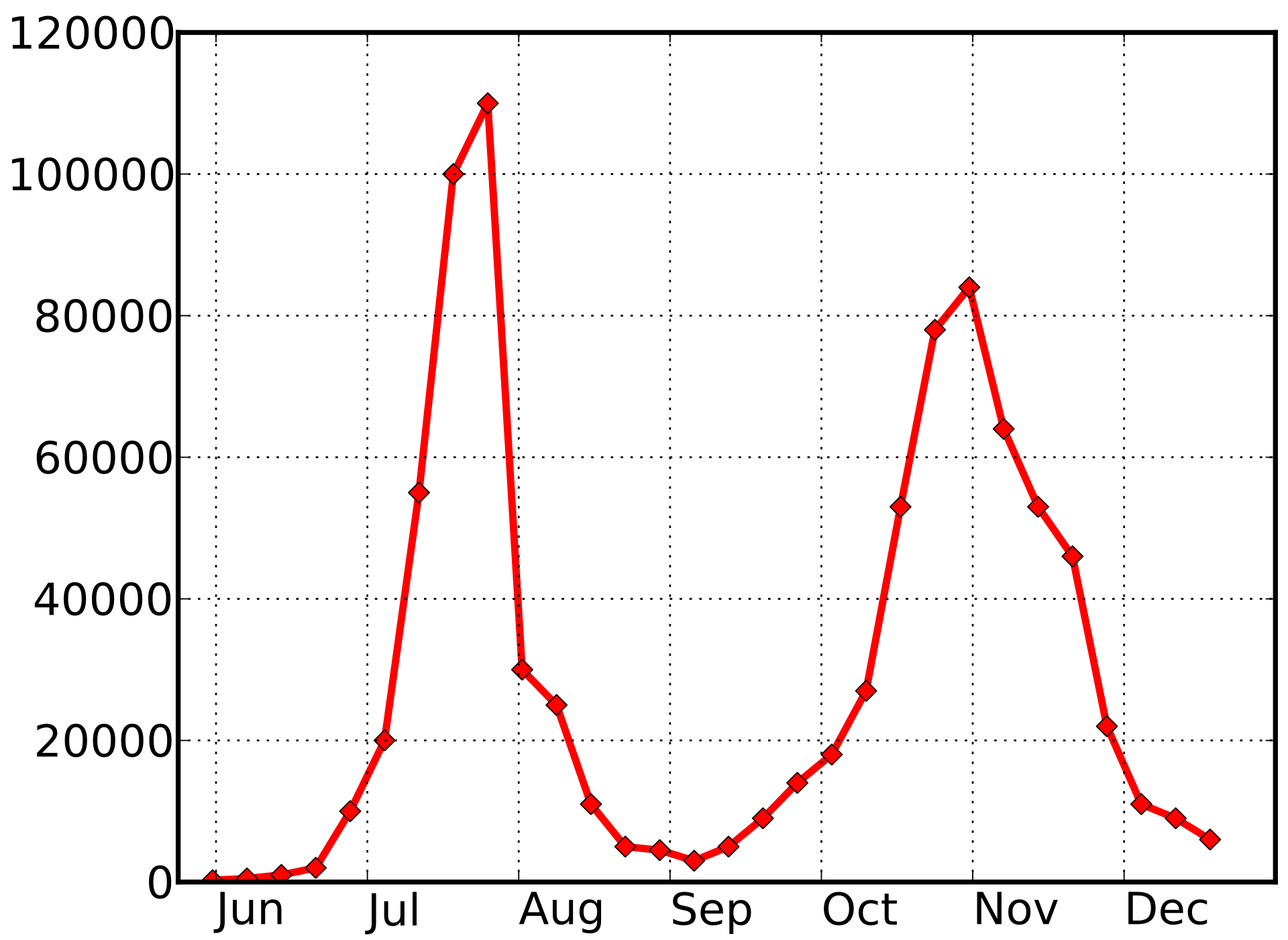
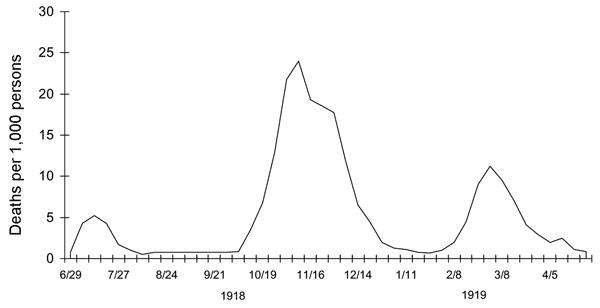
1918 influenza --- UK
 by Trevor Bedford
by Trevor Bedford
Human corona viruses have pronounced seasonal prevalence (Sweden)
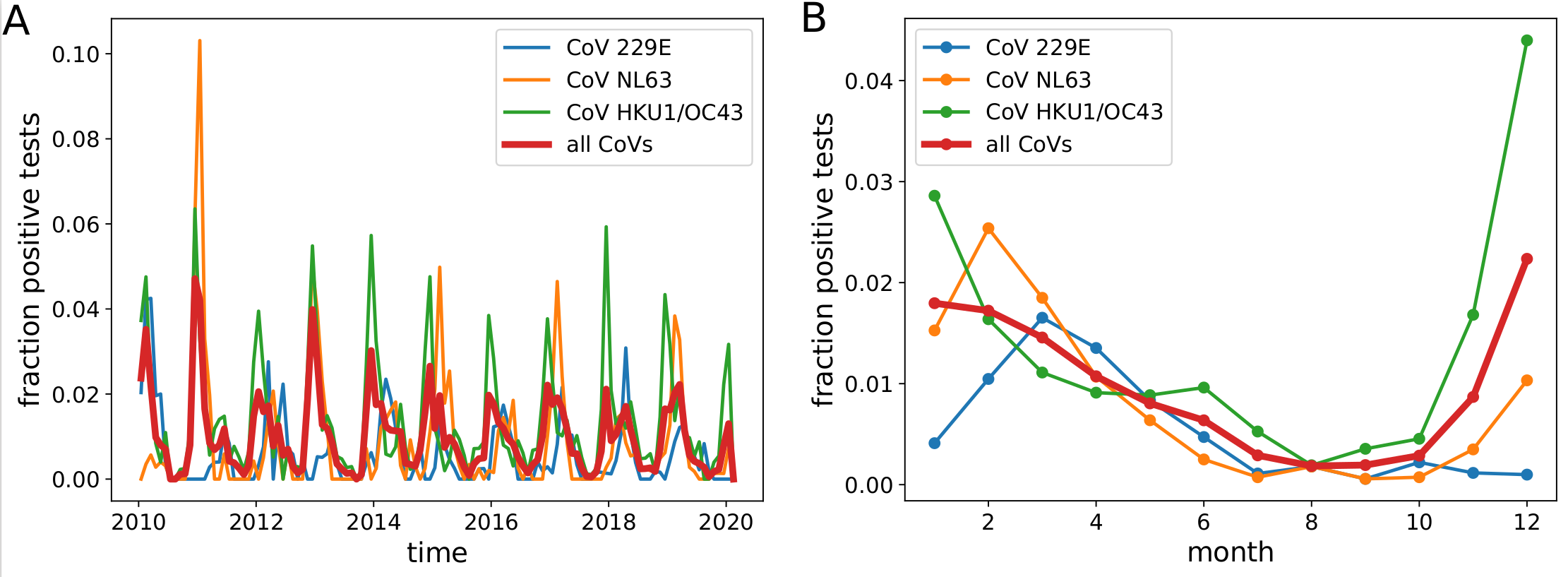
- Respiratory virus incl seasonal CoVs show seasonality
- Forcing through human behavior (indoor/outdoor activities).
- Control of the virus might be harder in temperate winter
- Absolute effect of seasonality unknown
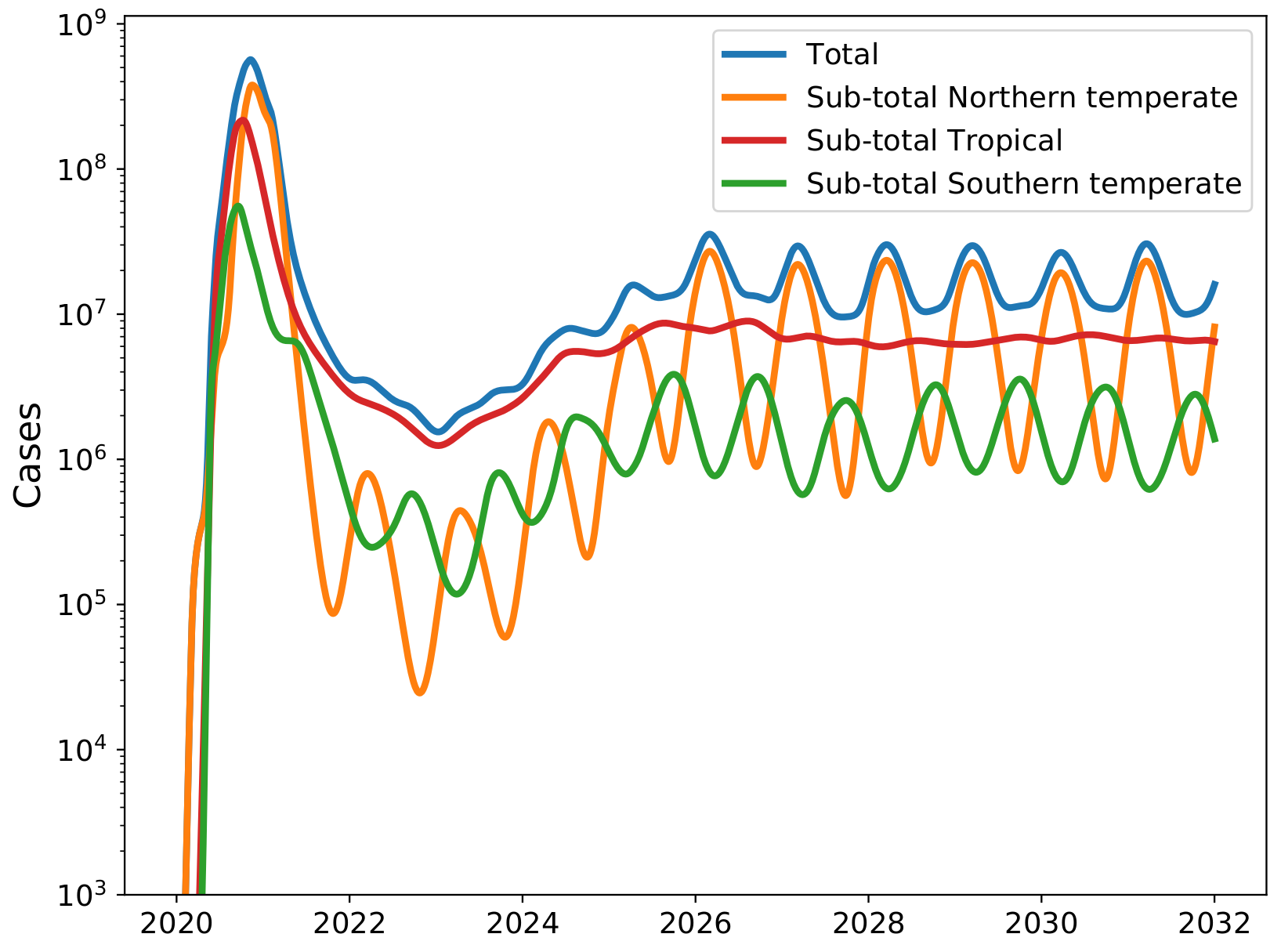
Potential transition to an endemic seasonal virus
Acknowledgments
Trevor Bedford and his lab -- terrific collaboration since 2014

especially James Hadfield, Emma Hodcroft, Ivan Aksamentov, Moira Zuber, and Tom Sibley
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID


Acknowledgments
- Robert Dyrdak
- Jan Albert
- Valentin Druelle
- Emma Hodcroft

Fighting misinformation
Conspiracy theories and sensationalist research
WRONG:
- COVID-19 has been with us for much longer
Genetic diversity suggests recent emergence, respiratory samples from last year are PCR negative - Recombinant with HIV
debunked by Trevor Bedford - The virus is more/less aggressive now...
No genetic changes shared by a majority of viruses since March - Prevalence is high, the mortality is low
False positive rates are not accounted for correctly, unrepresentative study populations - Some strains are more severe than others
confounders not accounted for - many more....
Colorful trees are easily misinterpreted...