Molecular epidemiology and phylogenetic analysis
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/202009_ECCVID_basics.html
Acknowledgments
Trevor Bedford and his lab -- terrific collaboration since 2014

especially James Hadfield, Emma Hodcroft, Ivan Aksamentov, Moira Zuber, and Tom Sibley
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID



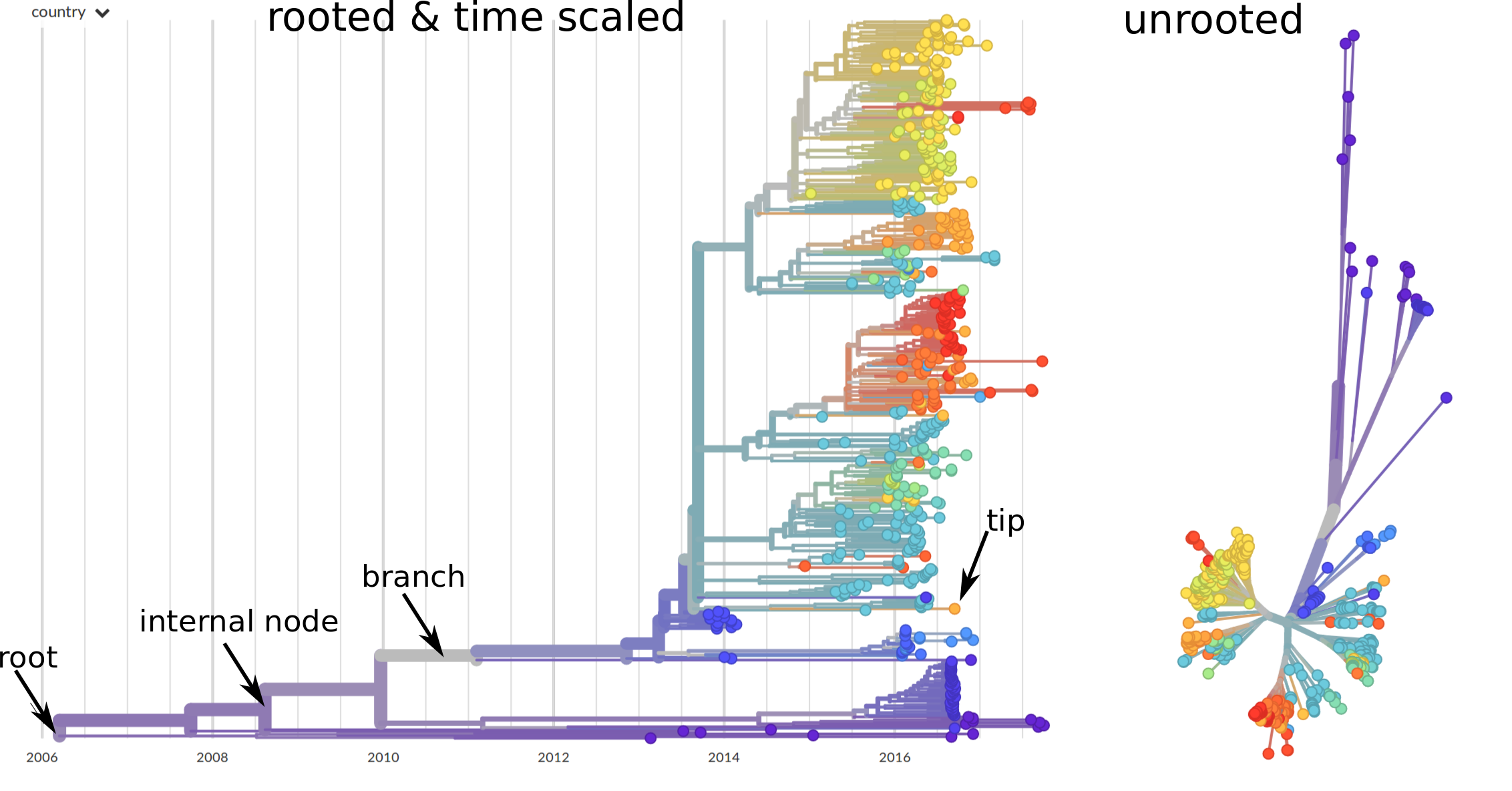
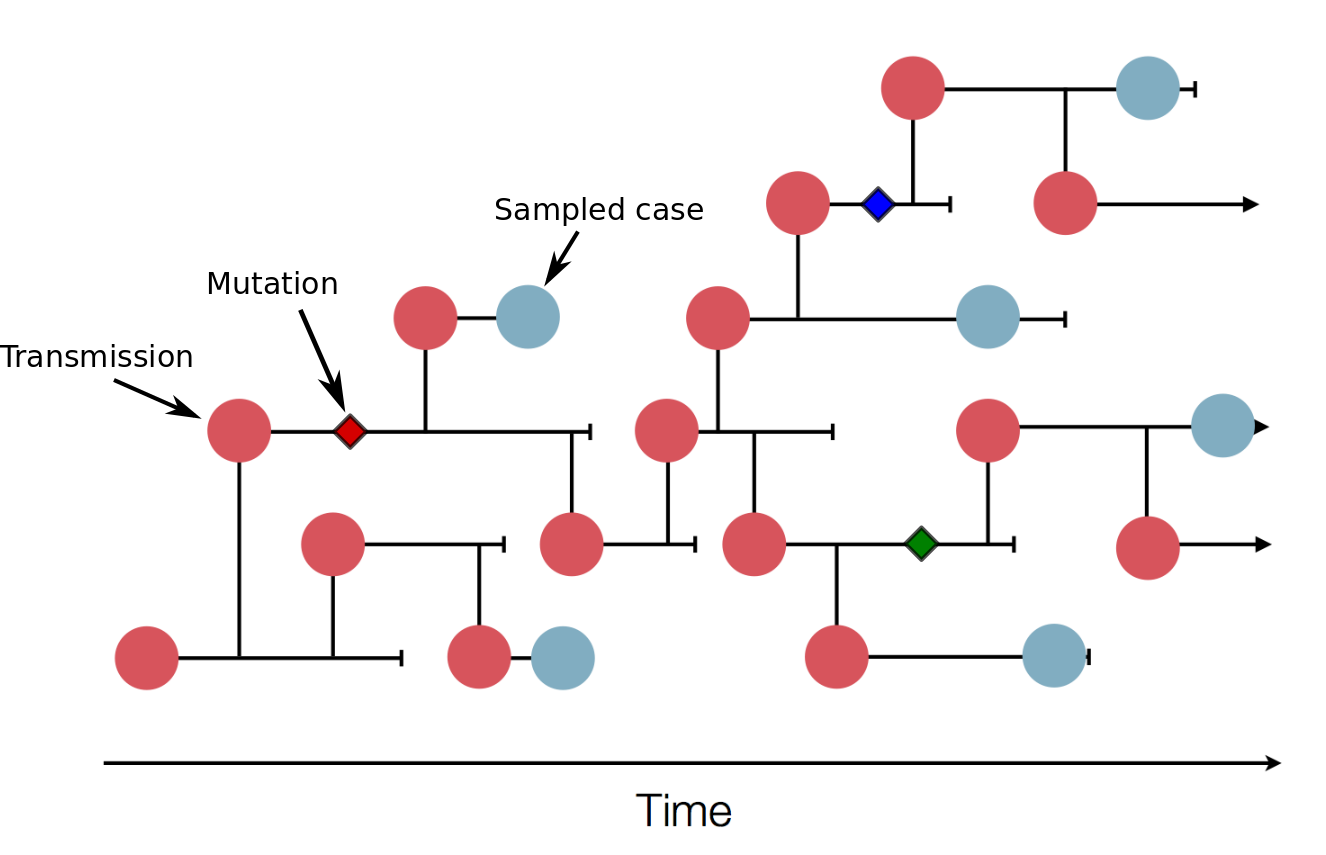
Sequences record the spread of pathogens




Mutations accumulate at a rate of $10^{-5}$ per site and day!
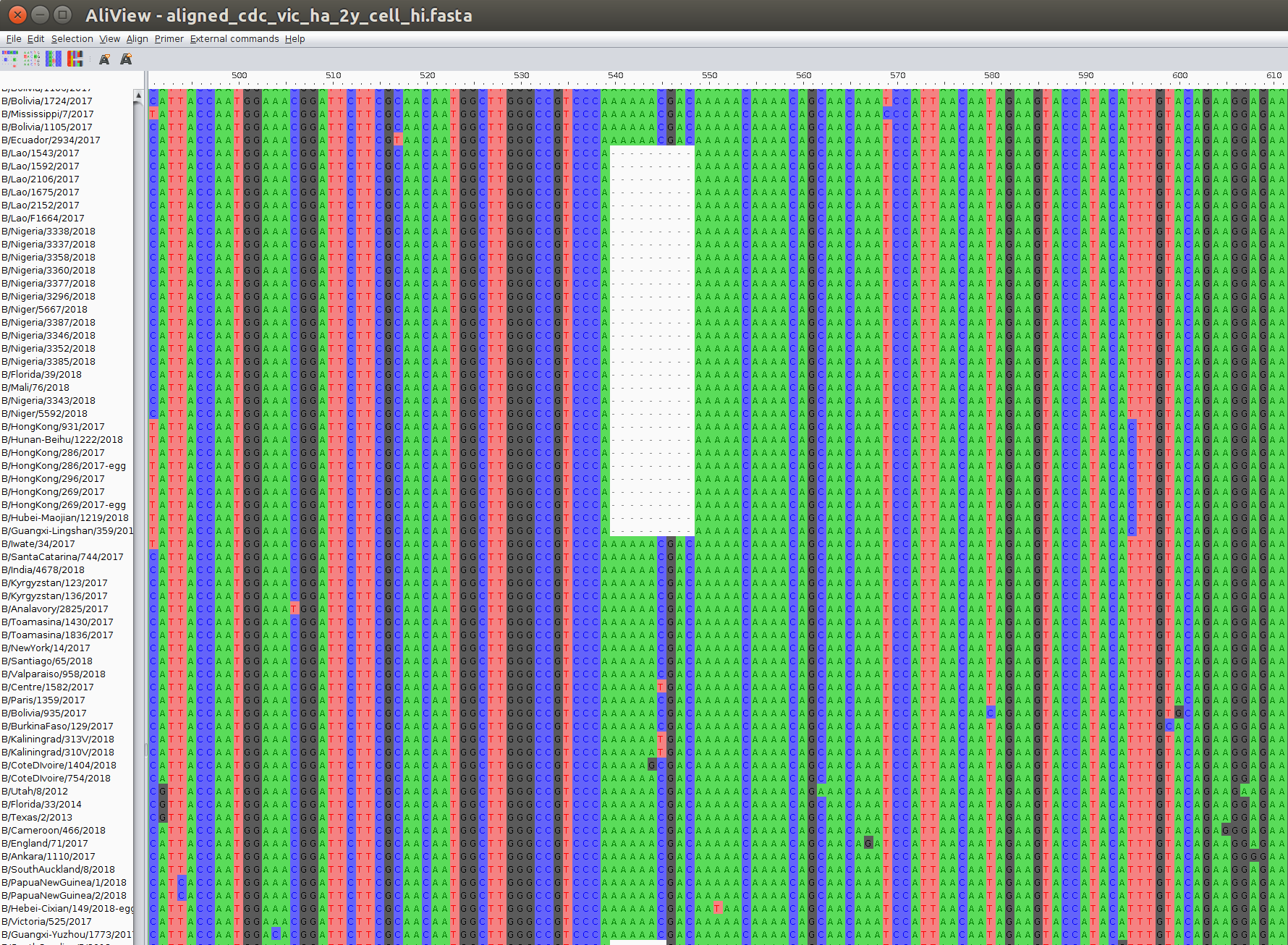
Sequences and Alignment
>SampleA
ACGCYTCGCGTCGACTCGCTAC...
>SampleB
CYTCGCCTCGACTCGCTACGCA...
>SampleC
GCYTCGCGTCGACTCGCTACGC...
>SampleC
GCYTCGCGTCGAGCTACGCATG...
>SampleA
ACGCYTCGCGTCGACTCGCTAC---...
>SampleB
---CYTCGCCTCGACTCGCTACGCA...
>SampleC
--GCYTCGCGTCGACTCGCTACGC-...
>SampleC
--GCYTCGCGTCGA---GCTACGCA...
Third party software: mafft, ClustalW, muscle...

Third party software: IQ-Tree, RAxML, fasttree...