Real-time tracking of SARS-CoV-2 spread and evolution
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/202009_deCOI.html
Acknowledgments
Trevor Bedford and his lab -- terrific collaboration since 2014

especially James Hadfield, Emma Hodcroft, Ivan Aksamentov, Moira Zuber, and Tom Sibley
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID


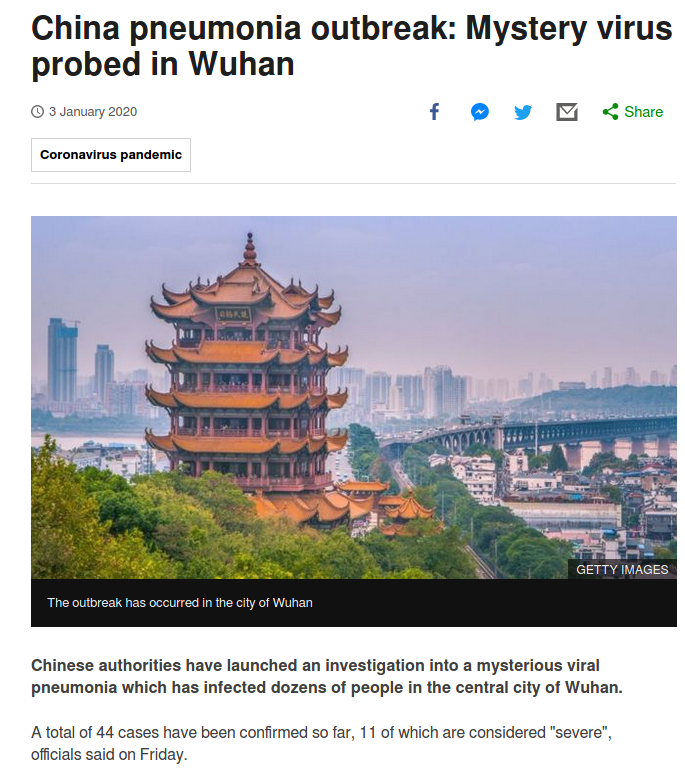 BBC
BBC
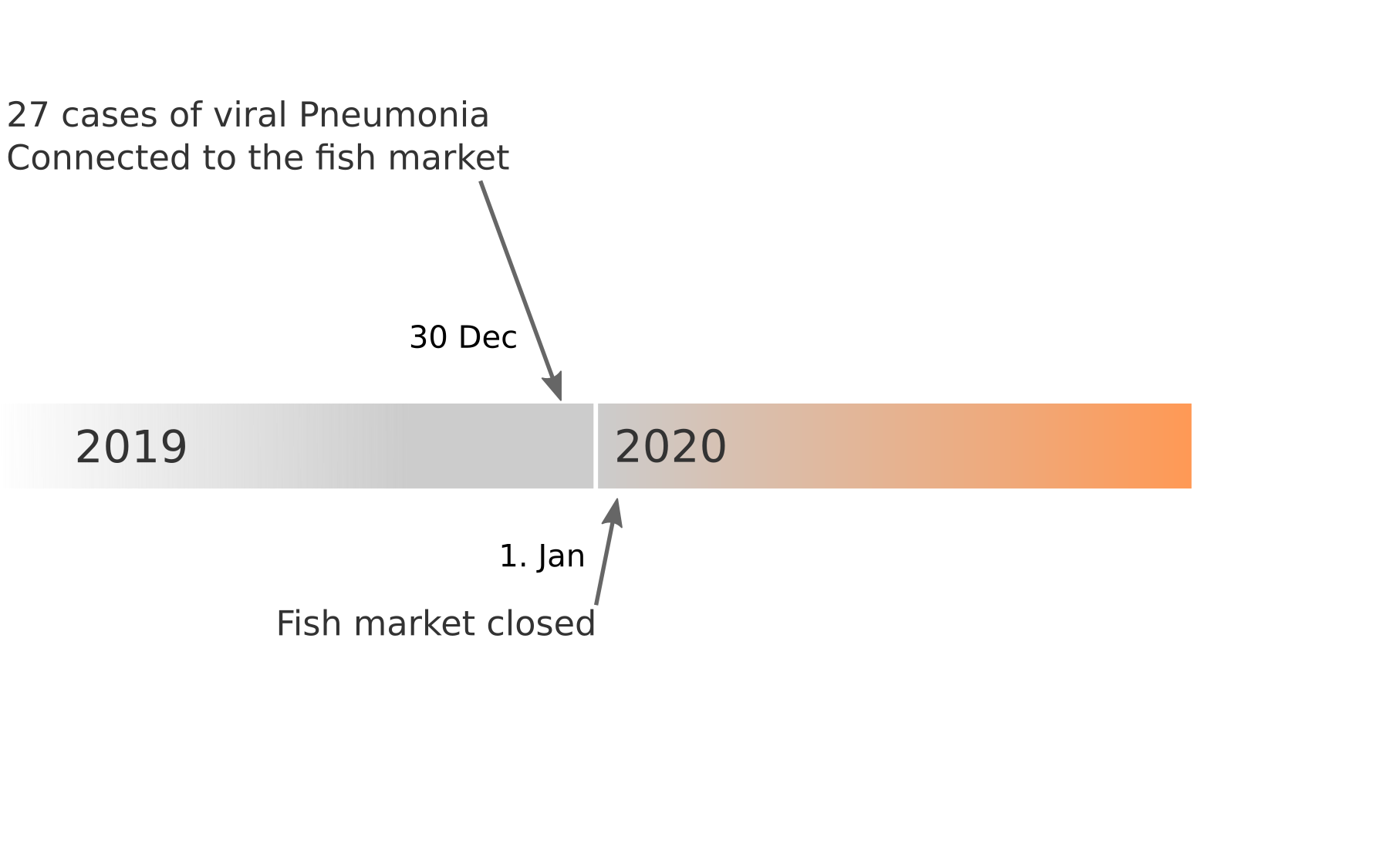 Data summarized by Ian MacKay
Data summarized by Ian MacKay
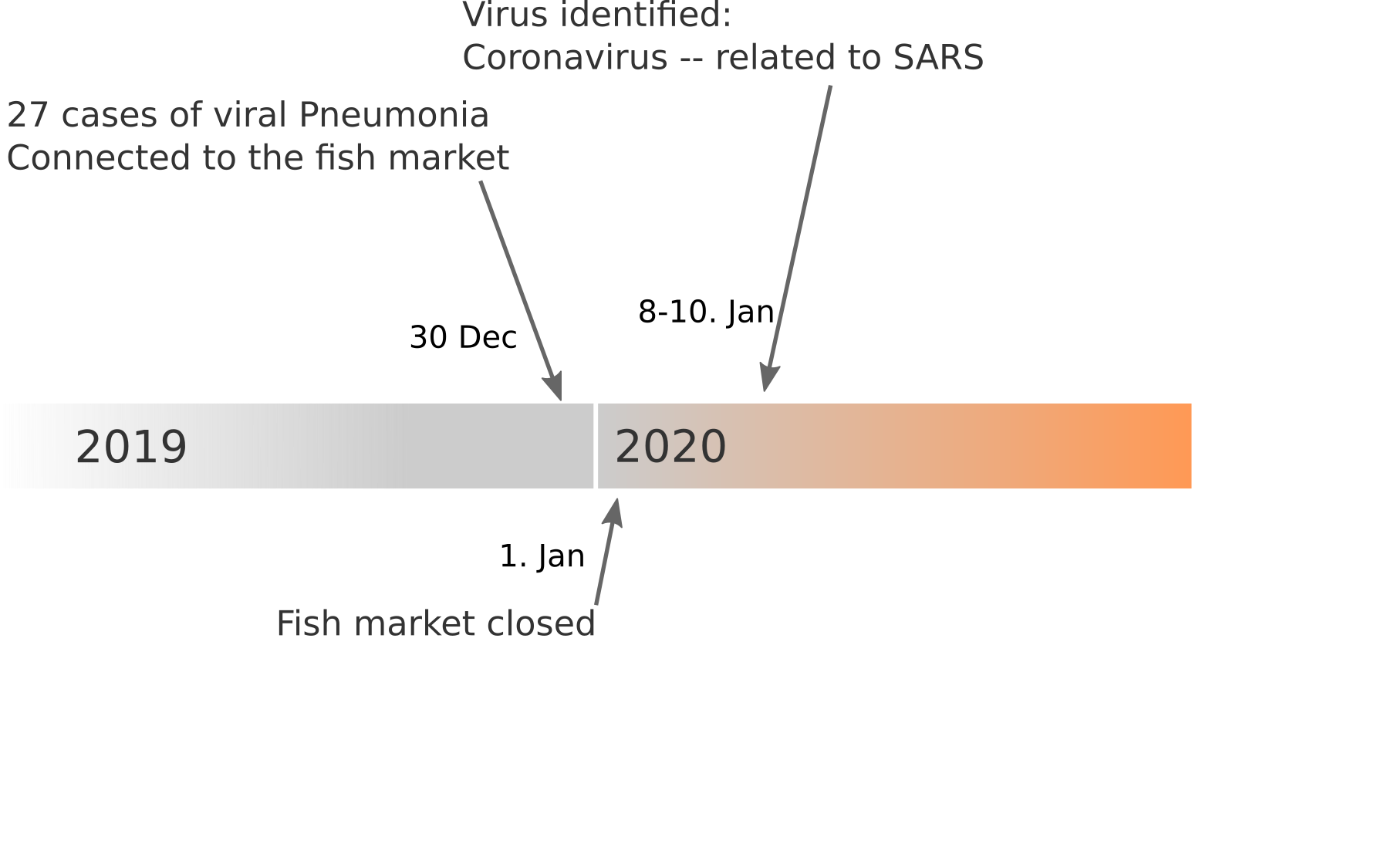 Data summarized by Ian MacKay
Data summarized by Ian MacKay
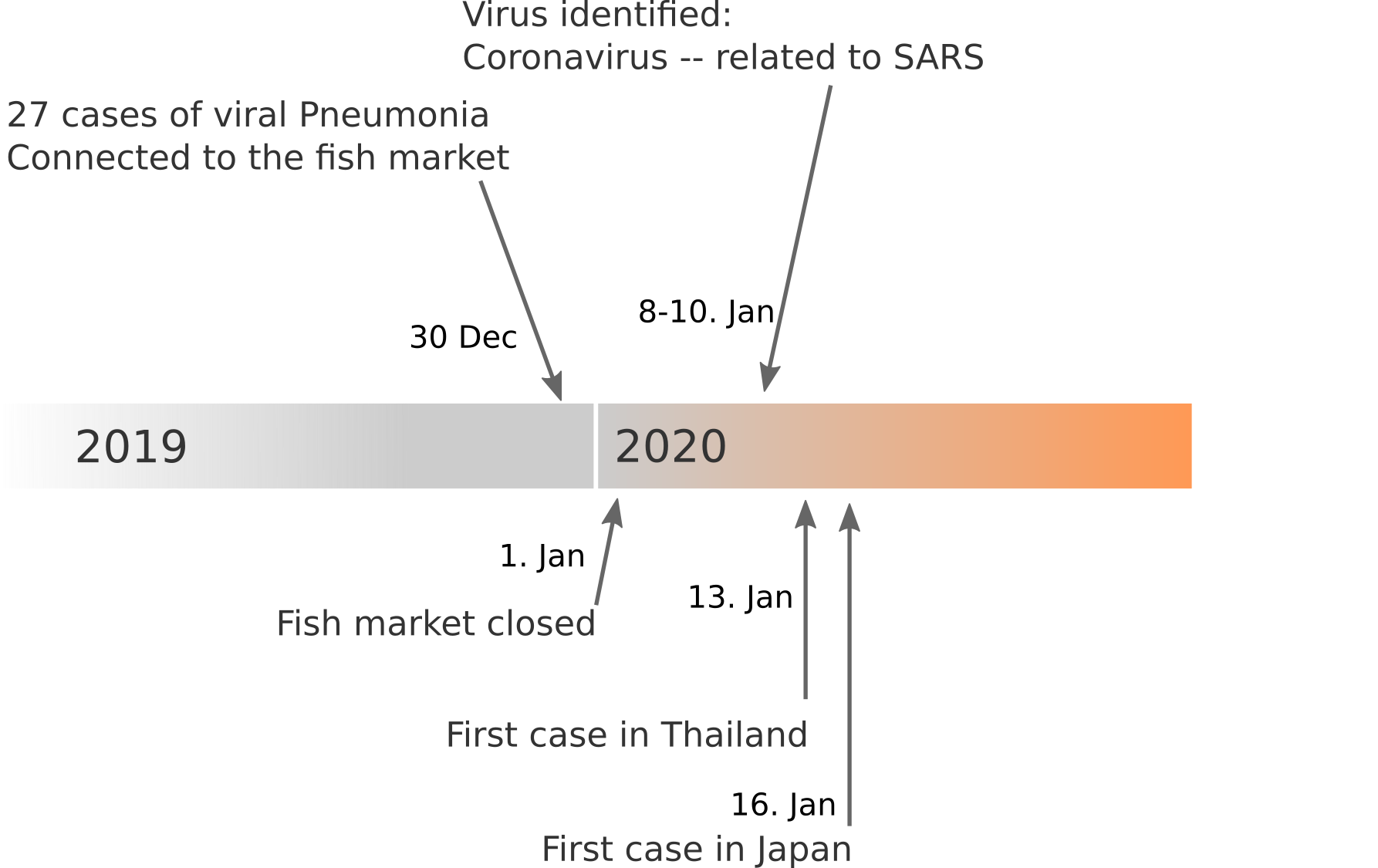 Data summarized by Ian MacKay
Data summarized by Ian MacKay
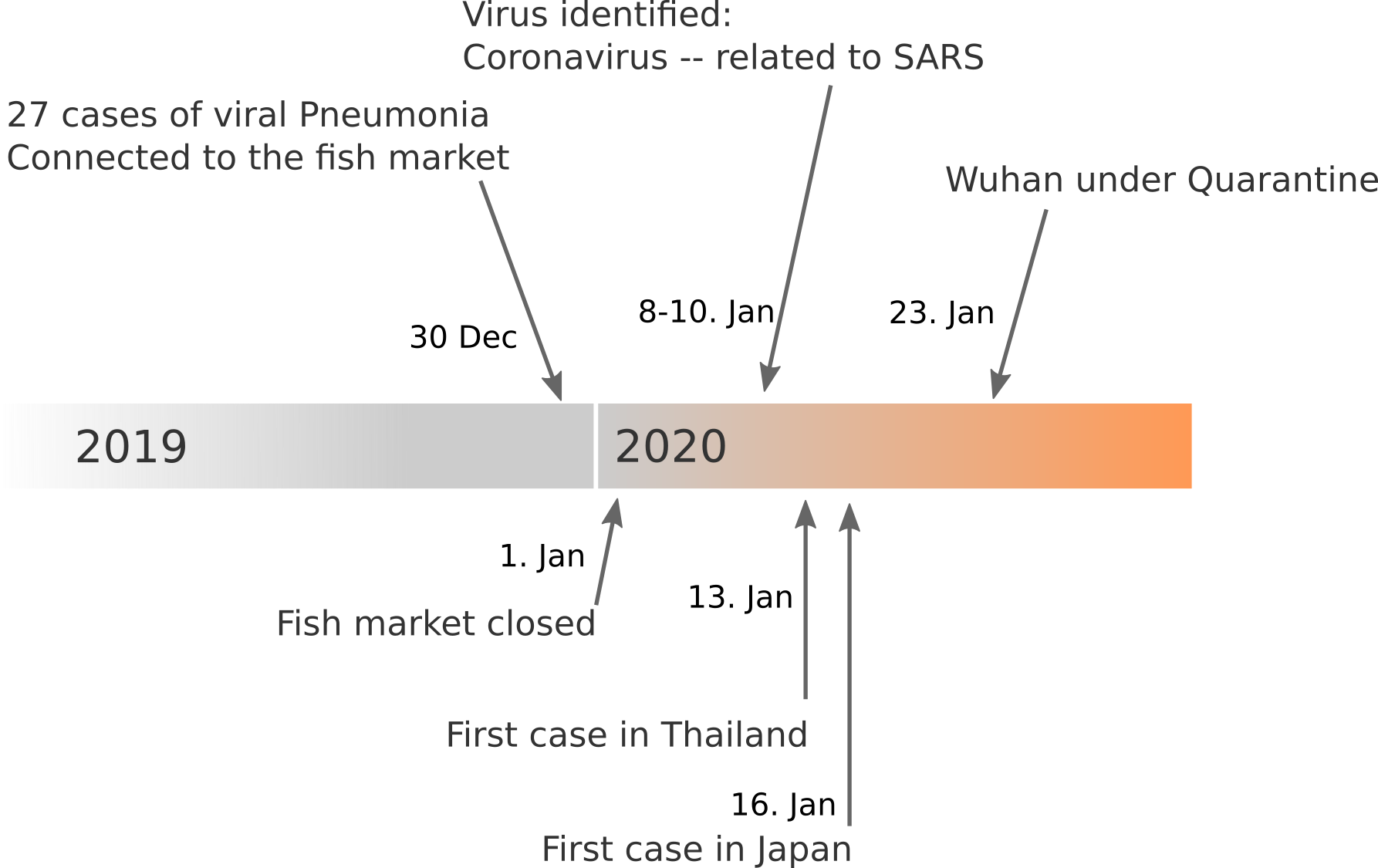 Data summarized by Ian MacKay
Data summarized by Ian MacKay
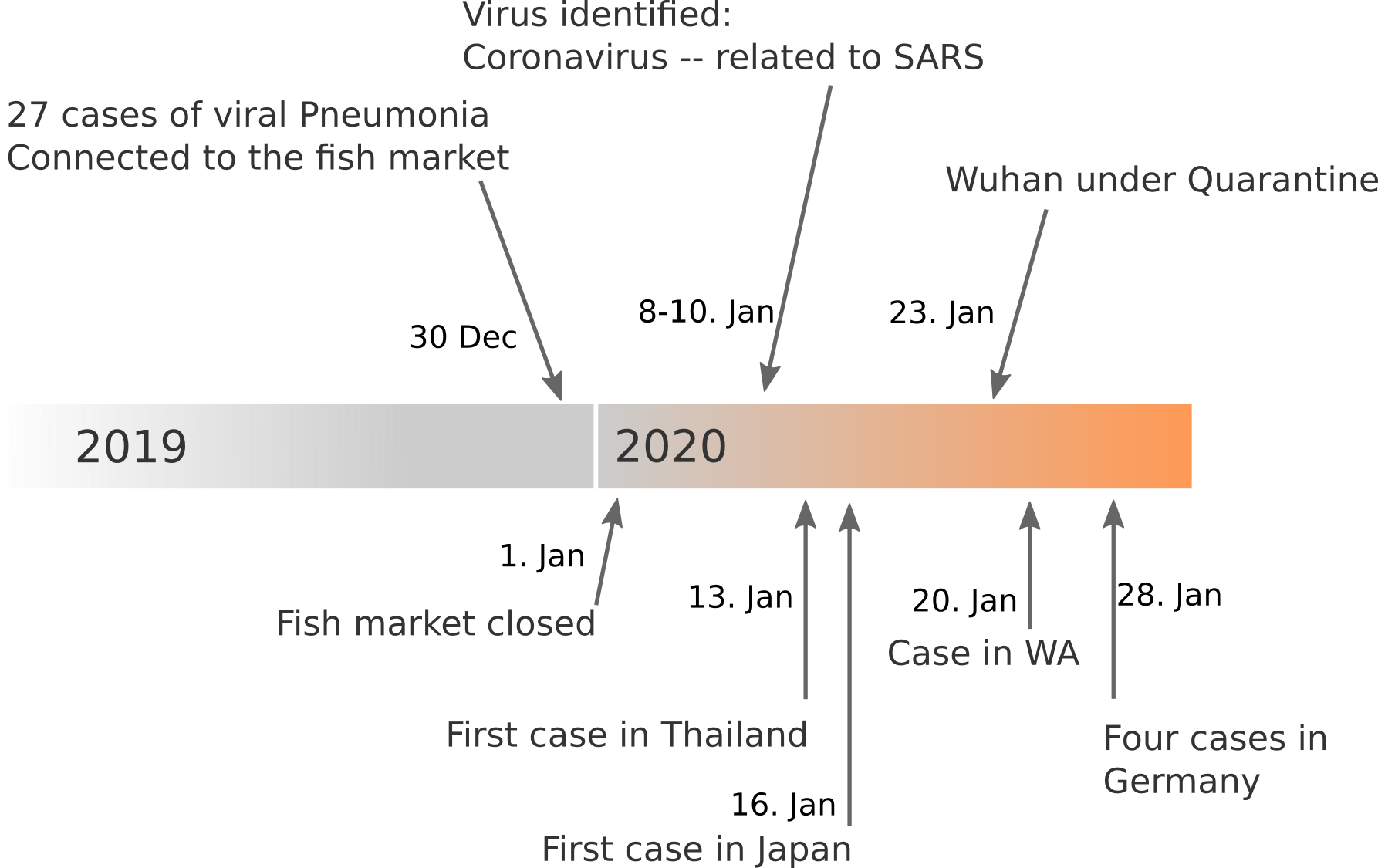 Data summarized by Ian MacKay
Data summarized by Ian MacKay
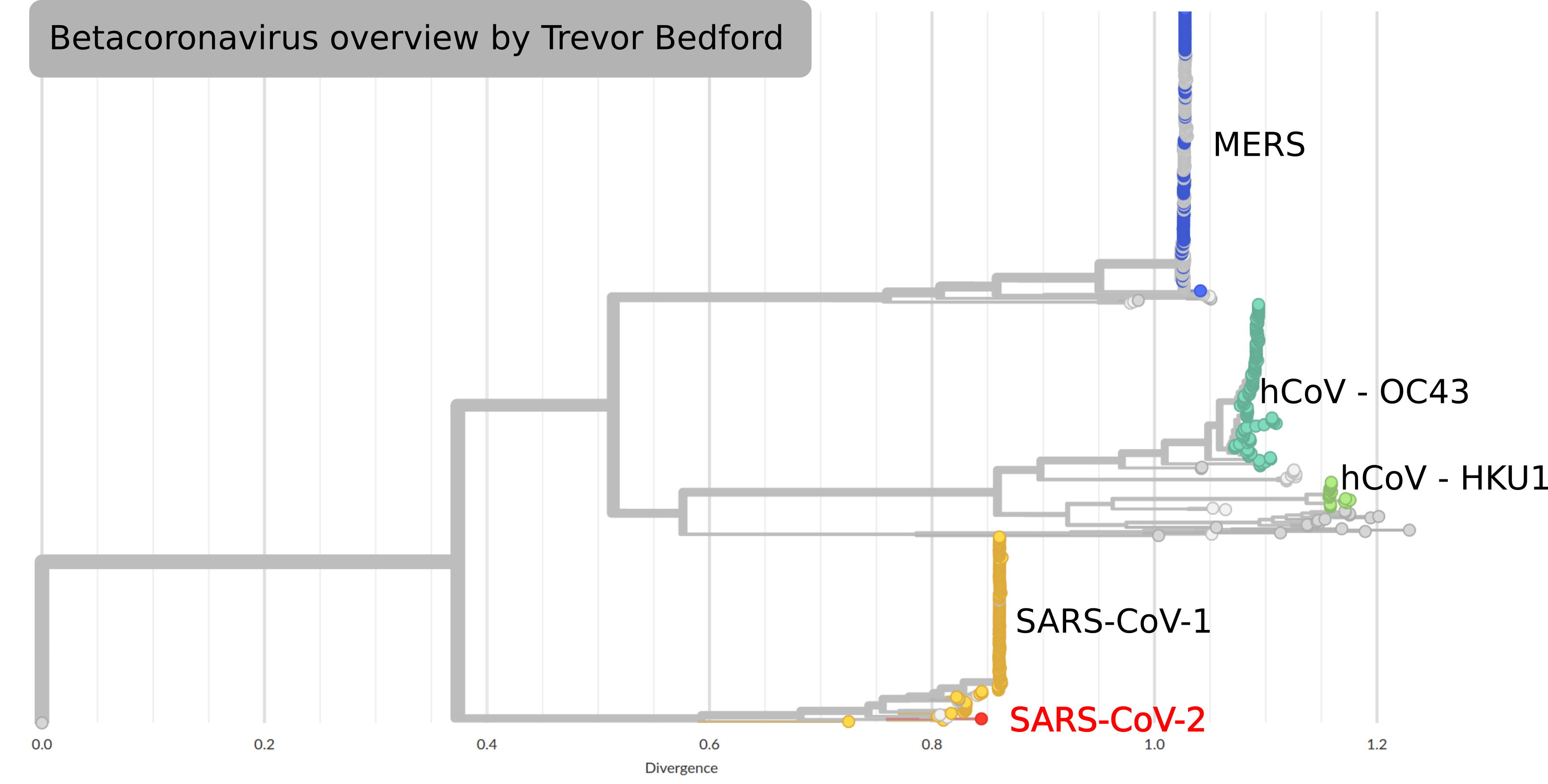 by Trevor Bedford
by Trevor Bedford
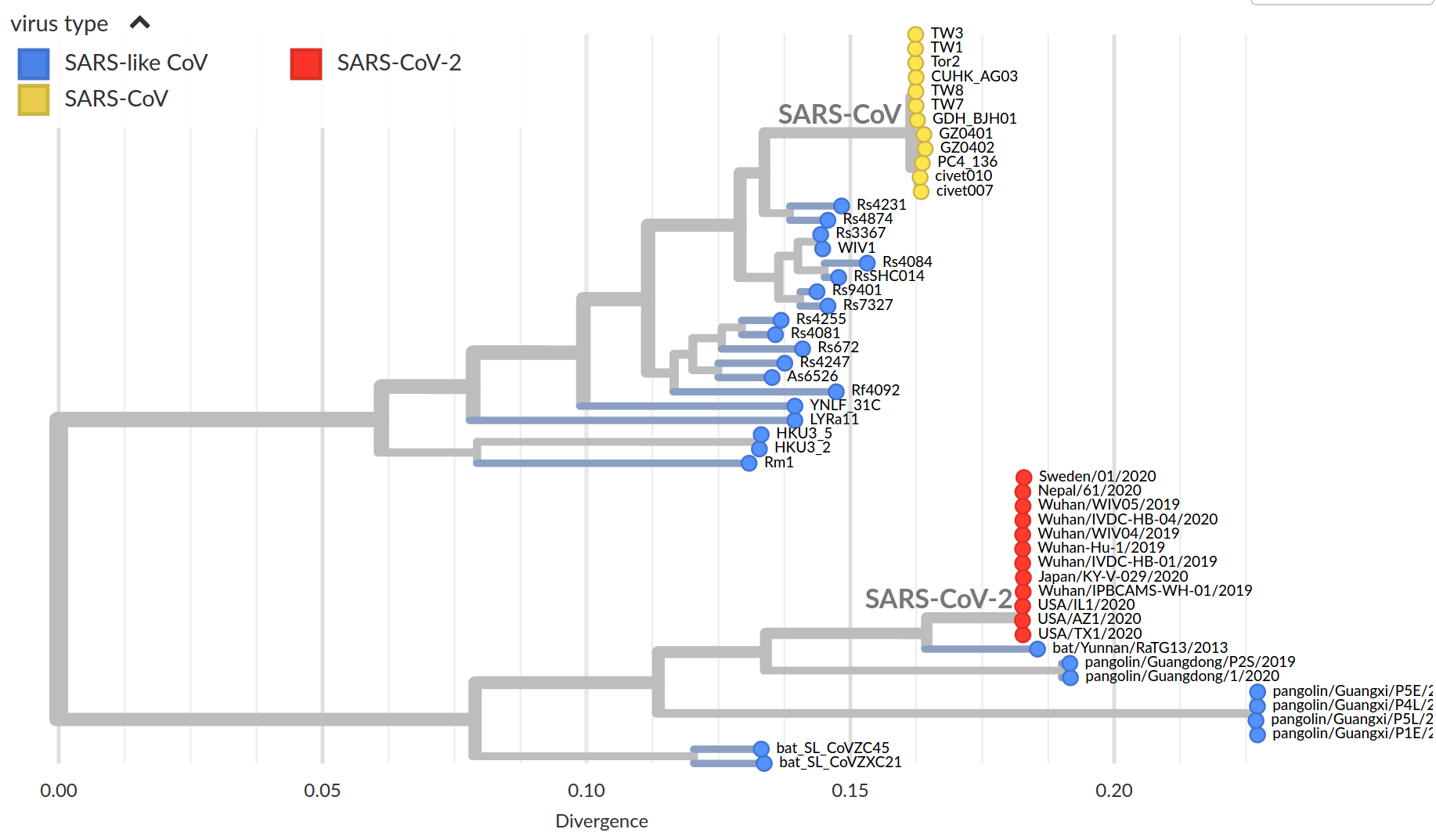 by Trevor Bedford
by Trevor Bedford
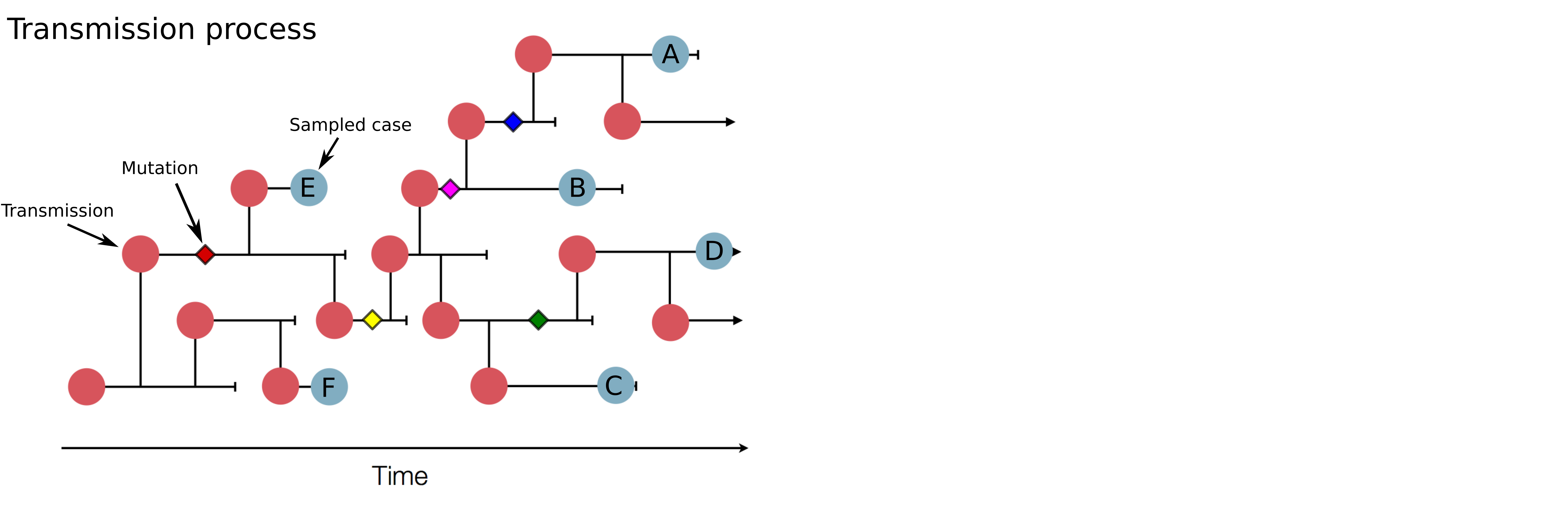
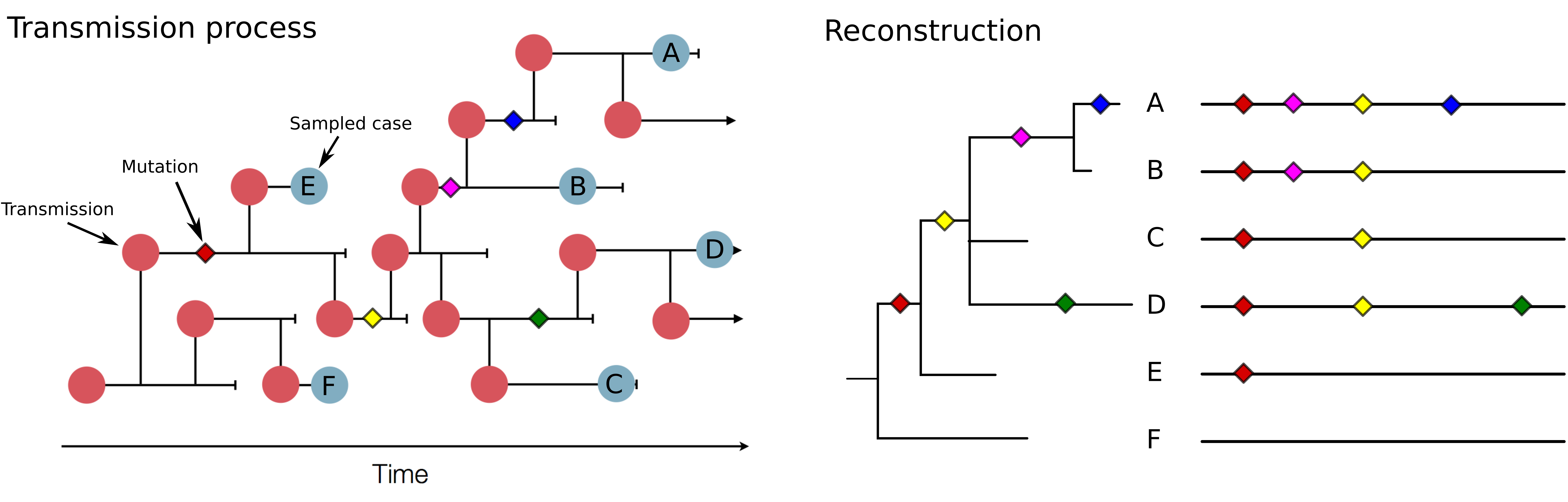
Tracking diversity and spread of SARS-CoV-2 in nextstrain
Available data on Jan 26
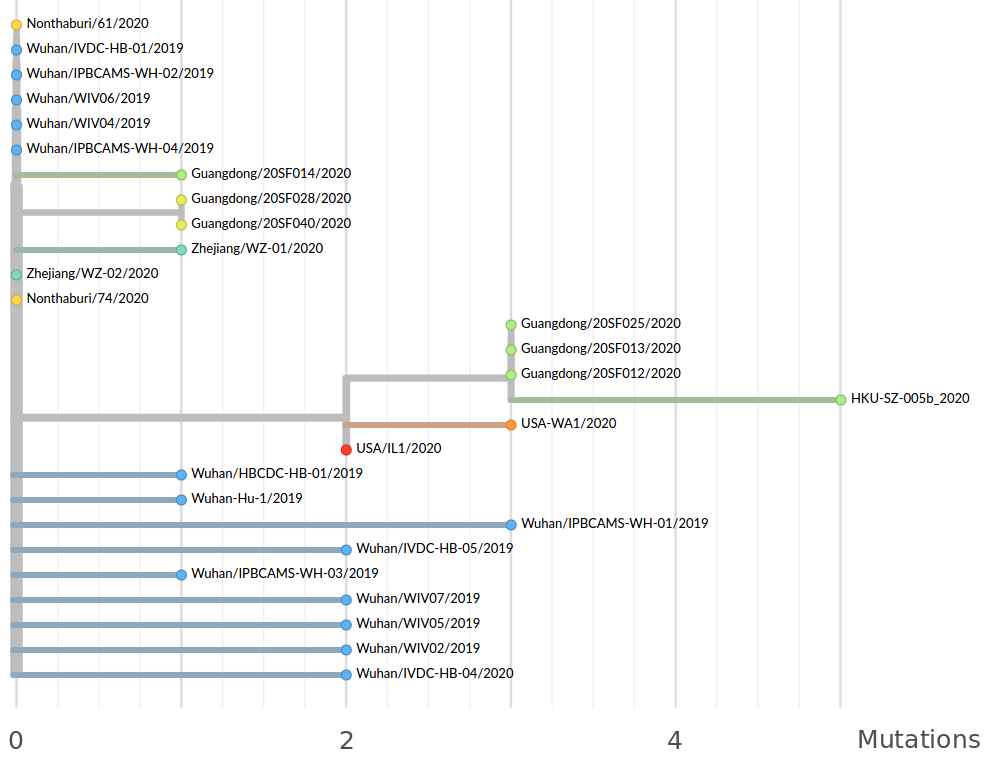
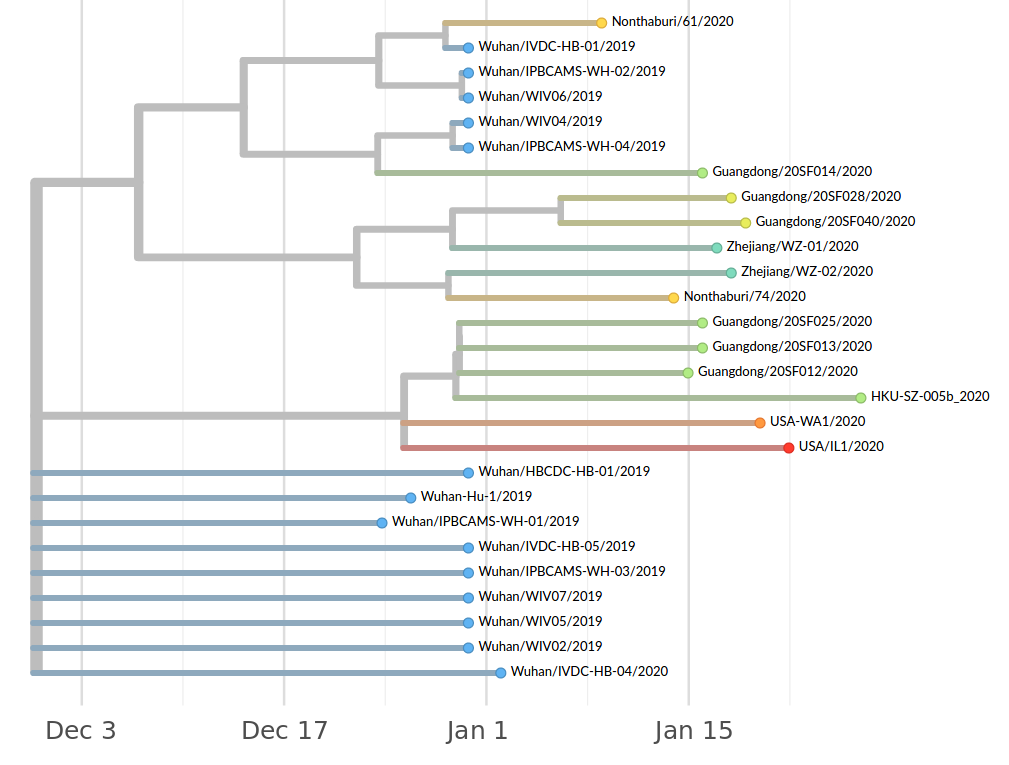
Early genomes differed by only a few mutations, suggesting very recent emergence
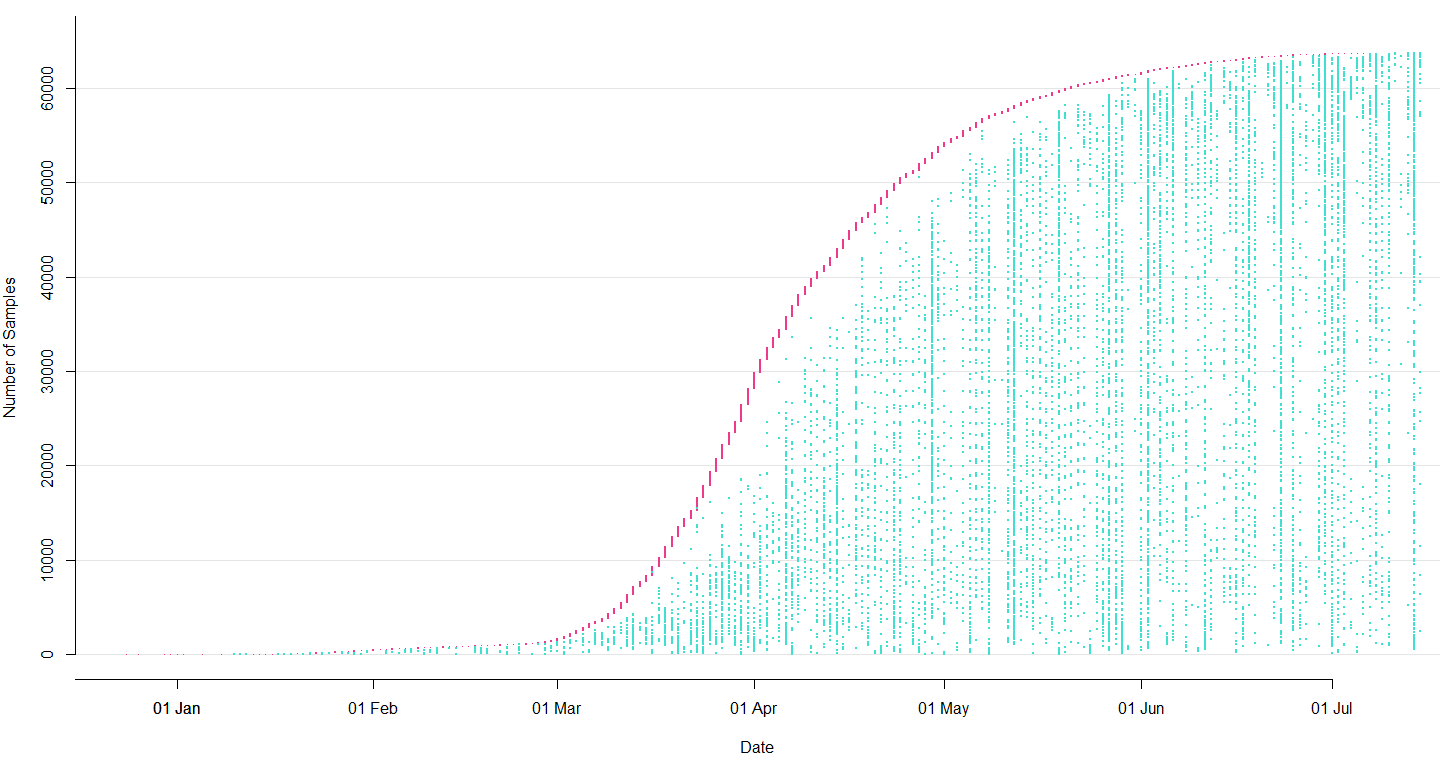
→ the closest to "real-time" we have experienced so far...
Figure by James Hadfield/Emma HodcroftSwiss sequences on August 31
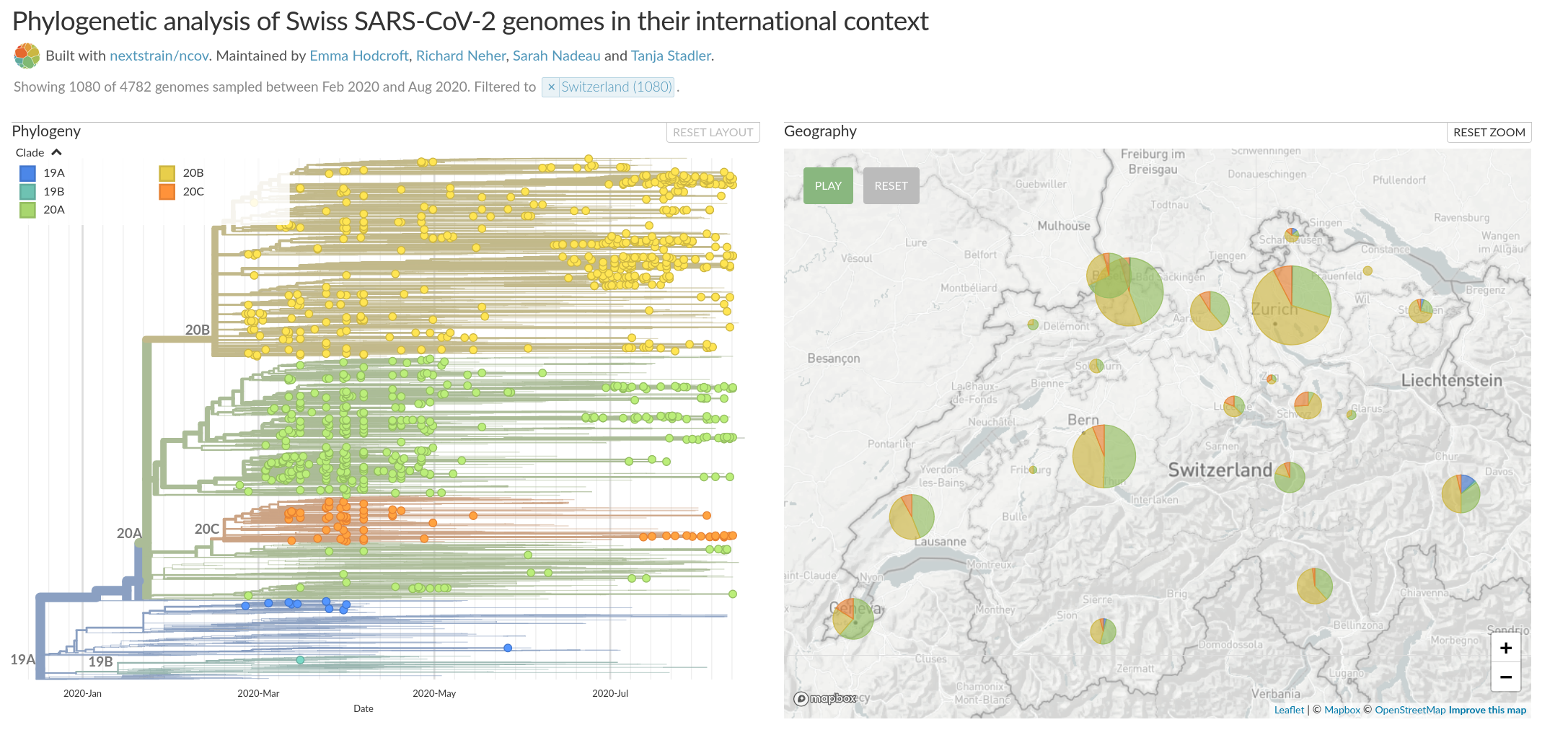
Genomic analysis as complement to contact tracing
A European cluster in summer 2020
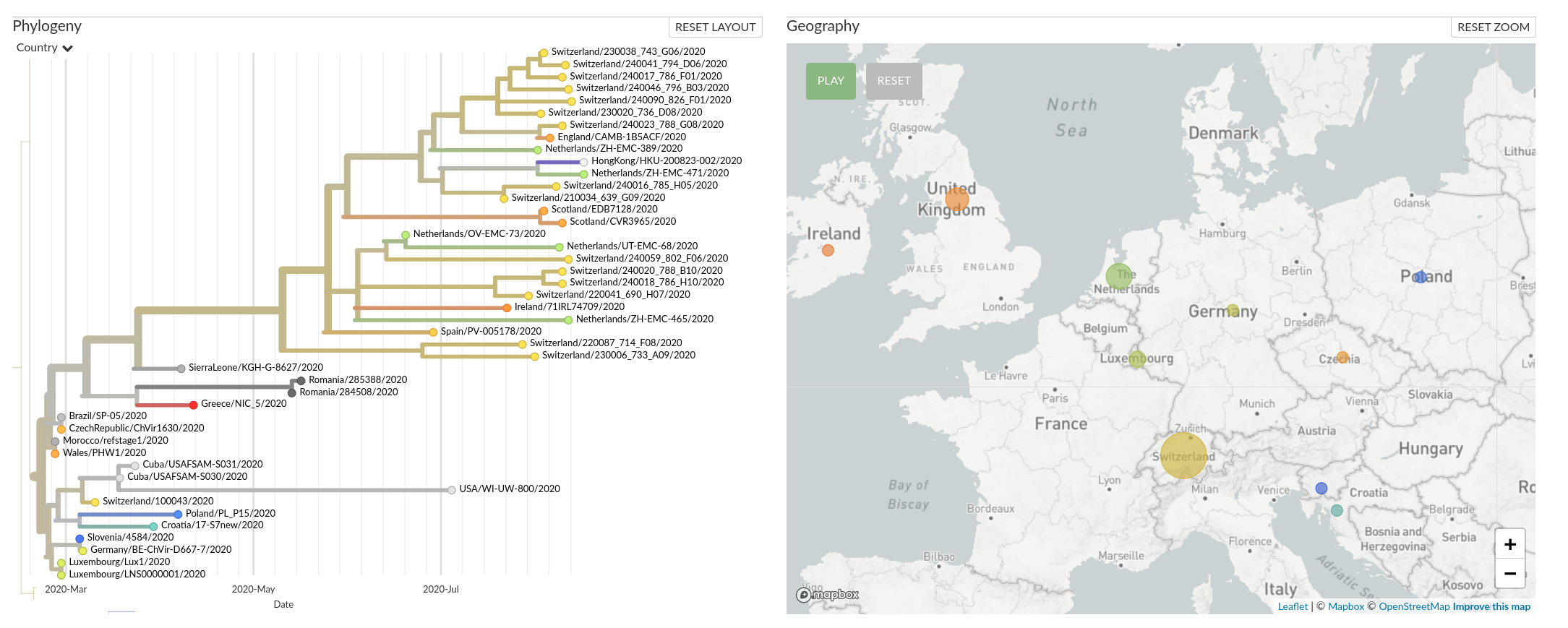
A European cluster in summer 2020
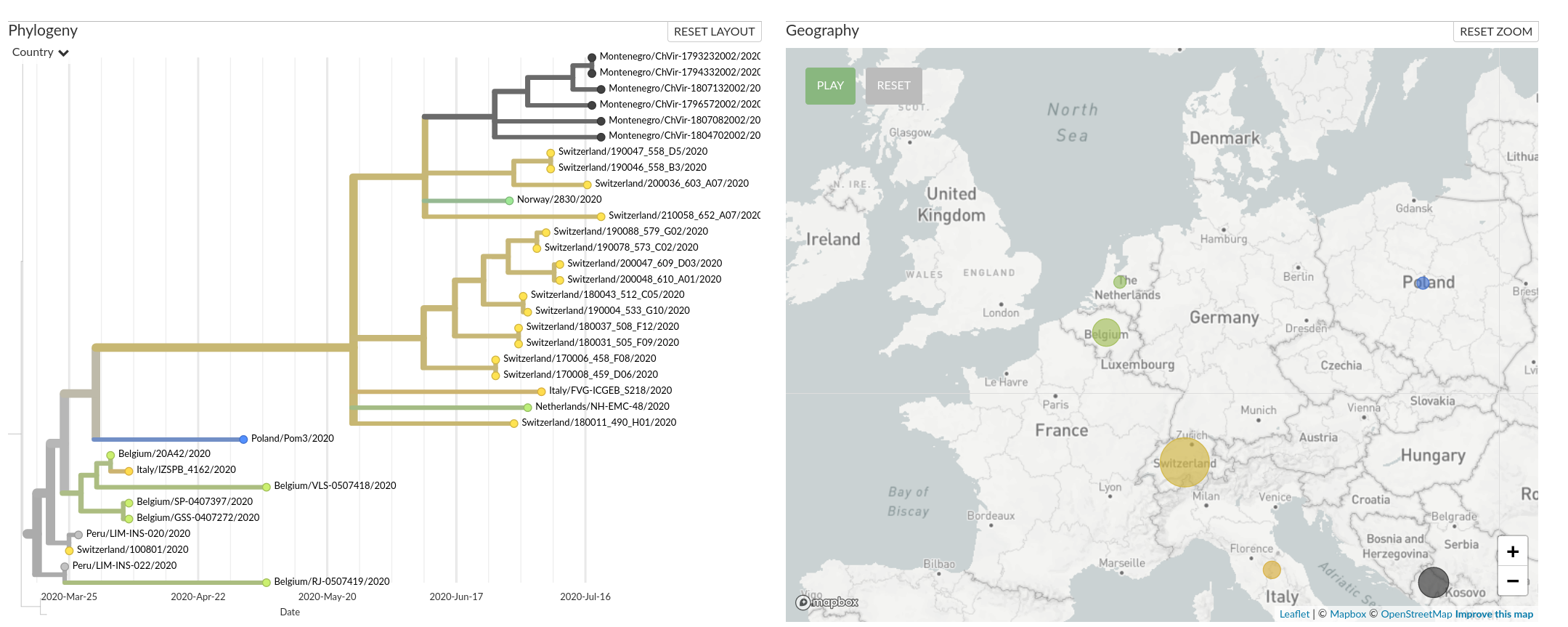
What's next?
- Social distancing is very effective
→ we can suppress the outbreak if we want! - Many Asian and European countries have successfully suppressed the virus while re-opening
- What role does seasonality have?
(indoor vs outdoor activities)
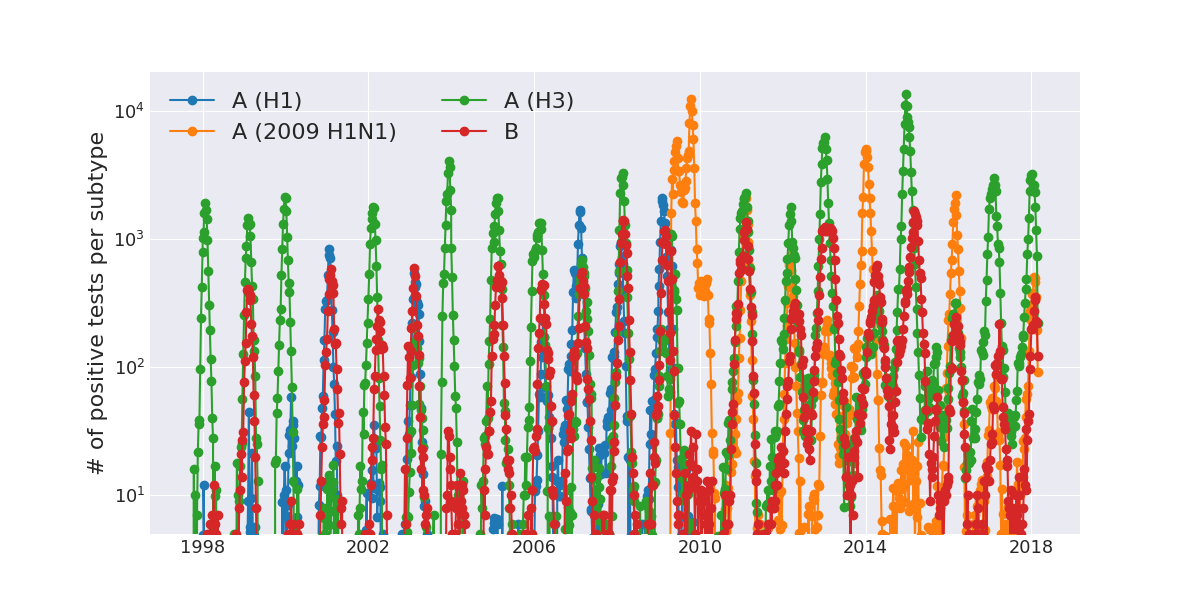
Seasonal incidence of influenza viruses
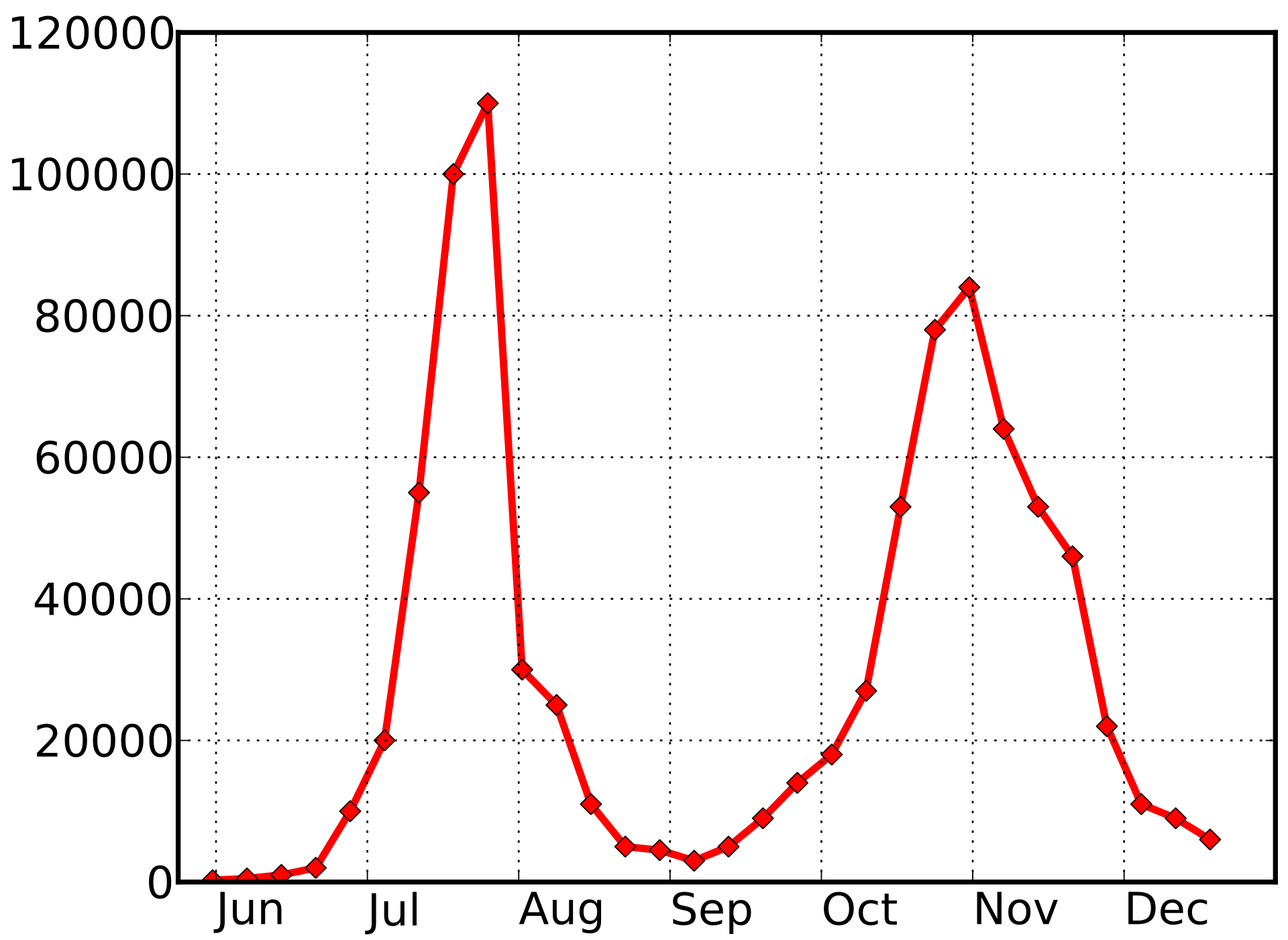
2009 pandemic influenza -- UK
Human corona viruses have pronounced seasonal prevalence (Sweden)
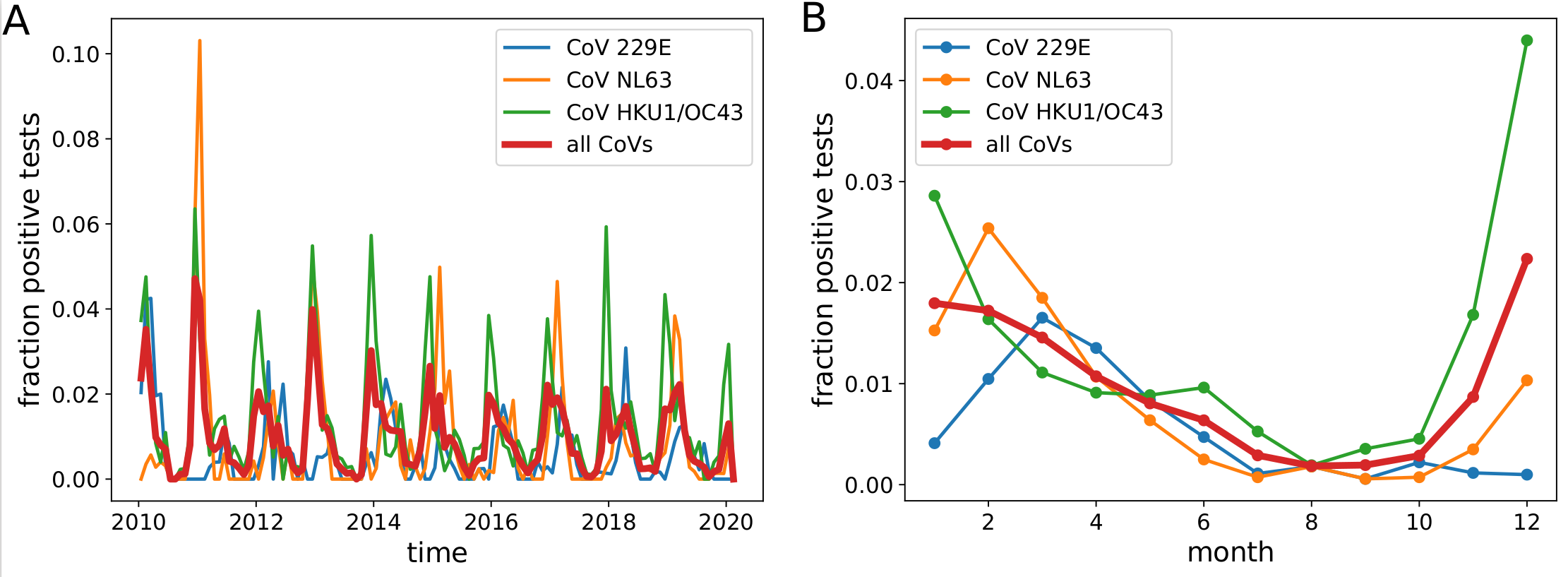
- Respiratory virus incl seasonal CoVs show seasonality
- Forcing through human behavior (indoor/outdoor activities).
- Control of the virus might be harder in temperate winter
- Absolute effect of seasonality unknown