Modeling and predicting RNA virus evolution
Richard Neher
Biozentrum & SIB, University of Basel
slides at neherlab.org/202010_KNAW_long.html
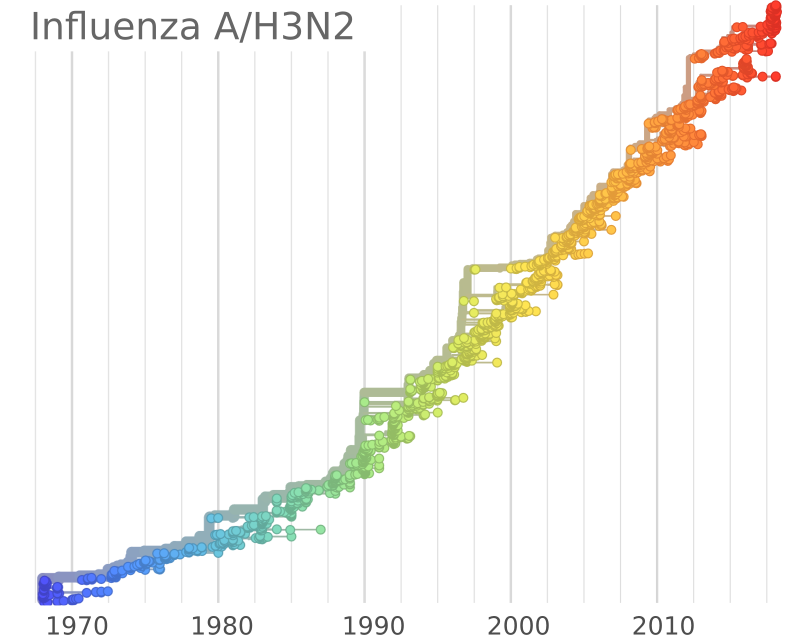
Human seasonal influenza viruses

Positive tests for influenza in the USA by week
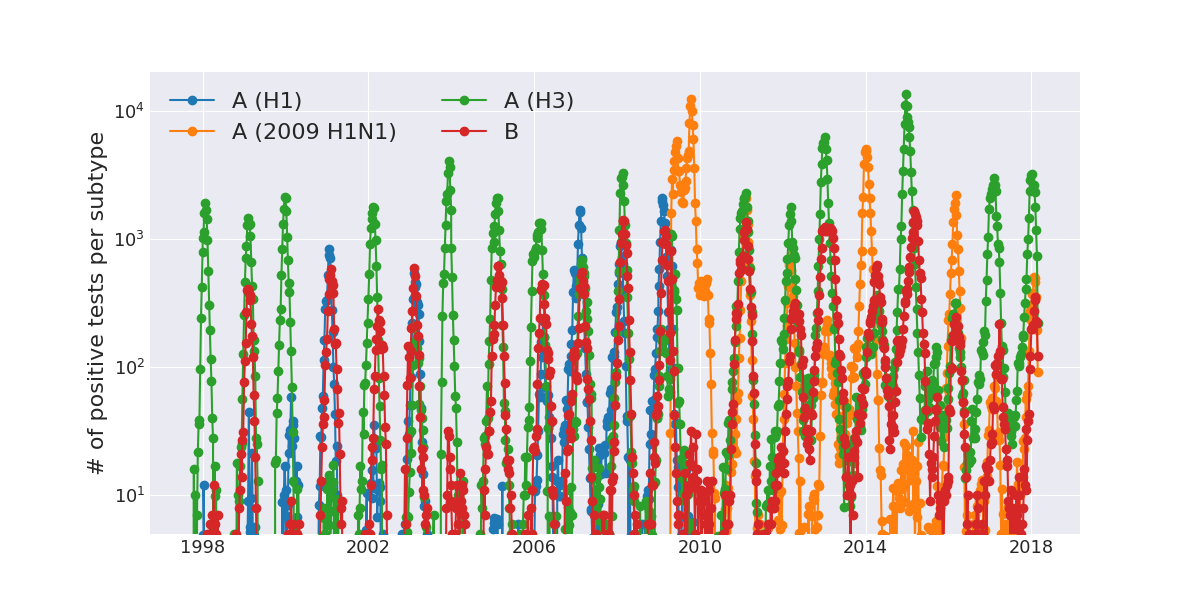 Data by the US CDC
Data by the US CDC



- Influenza viruses evolve to avoid human immunity
- Vaccines need frequent updates

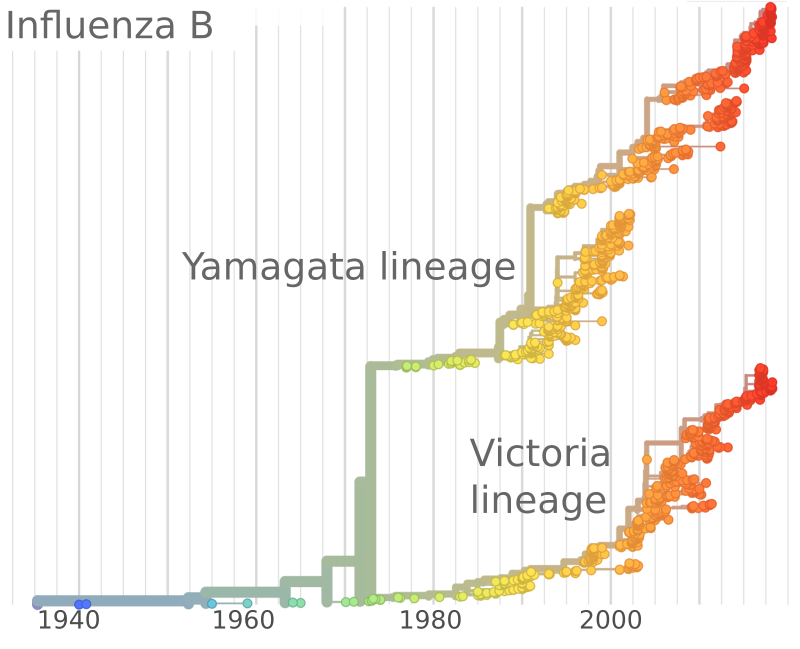
Influenza B viruses have split into two lineages
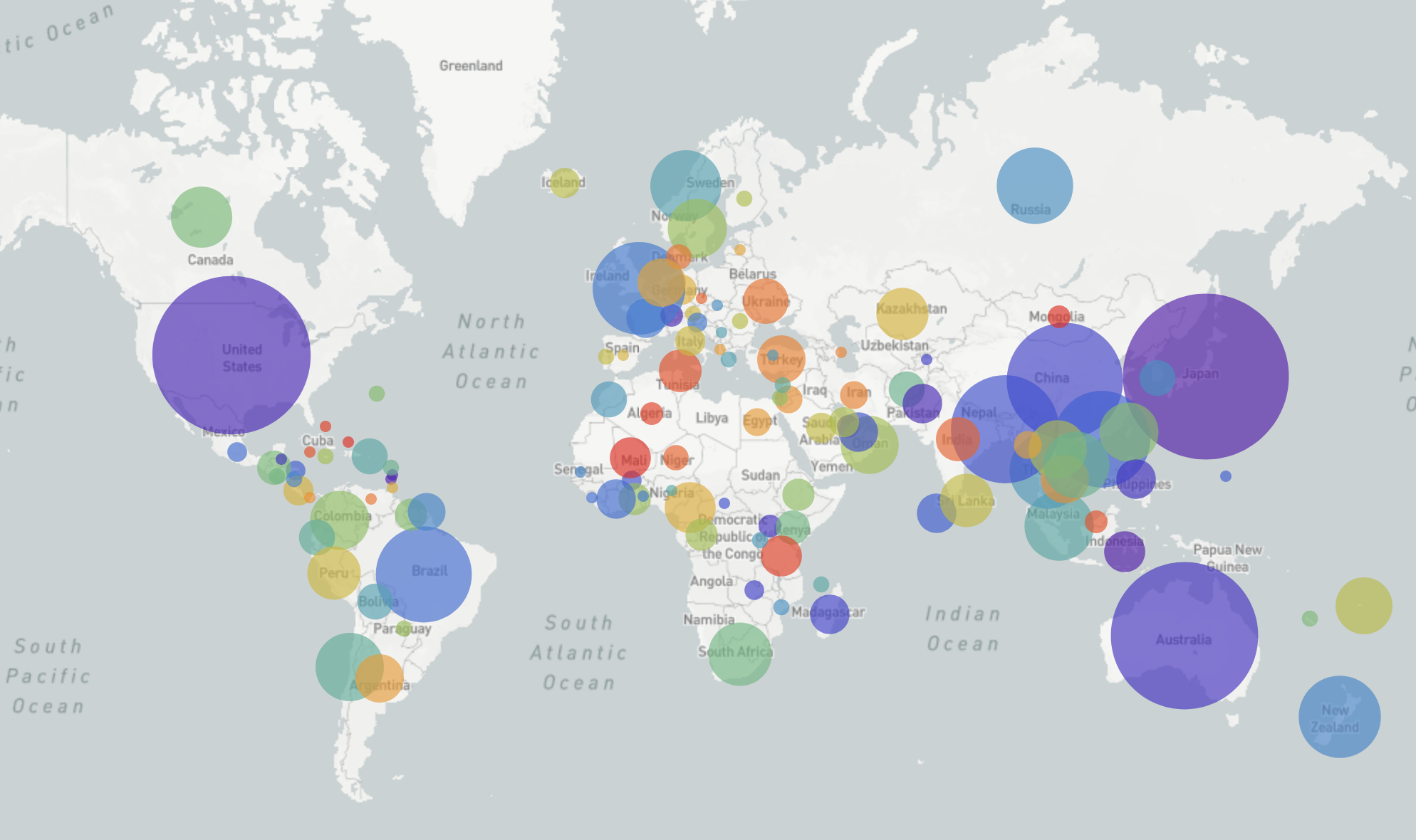
GISRS and GISAID -- Influenza virus surveillance
- comprehensive coverage of the world
- timely sharing of data through GISAID -- often within 2-3 weeks of sampling
- hundreds of sequences per week (in peak months)
→ requires continuous analysis and easy dissemination
→ interpretable and intuitive visualization
nextflu.org
joint project with Trevor Bedford & his lab
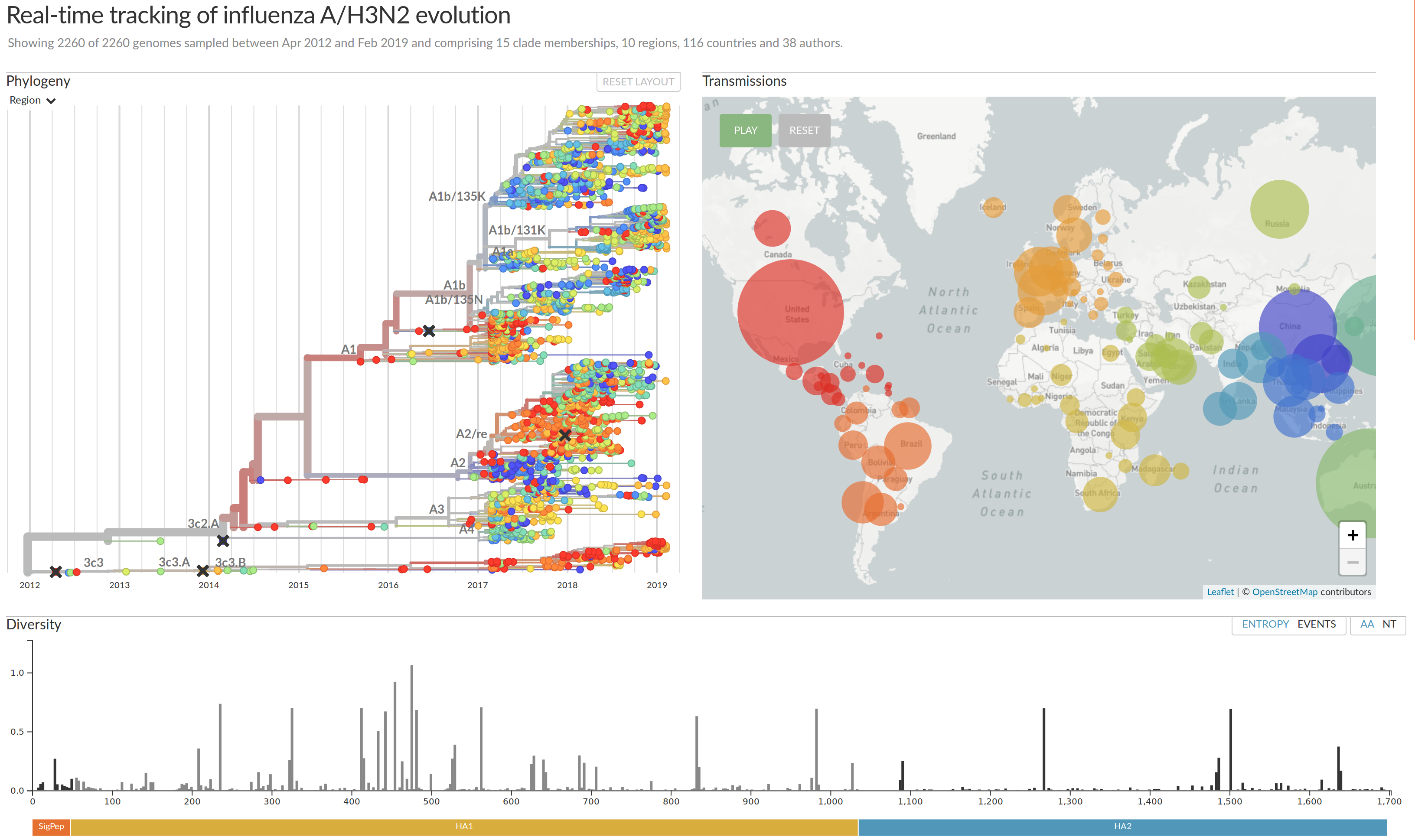
Beyond tracking: can we predict?
Fitness variation in rapidly adapting populations
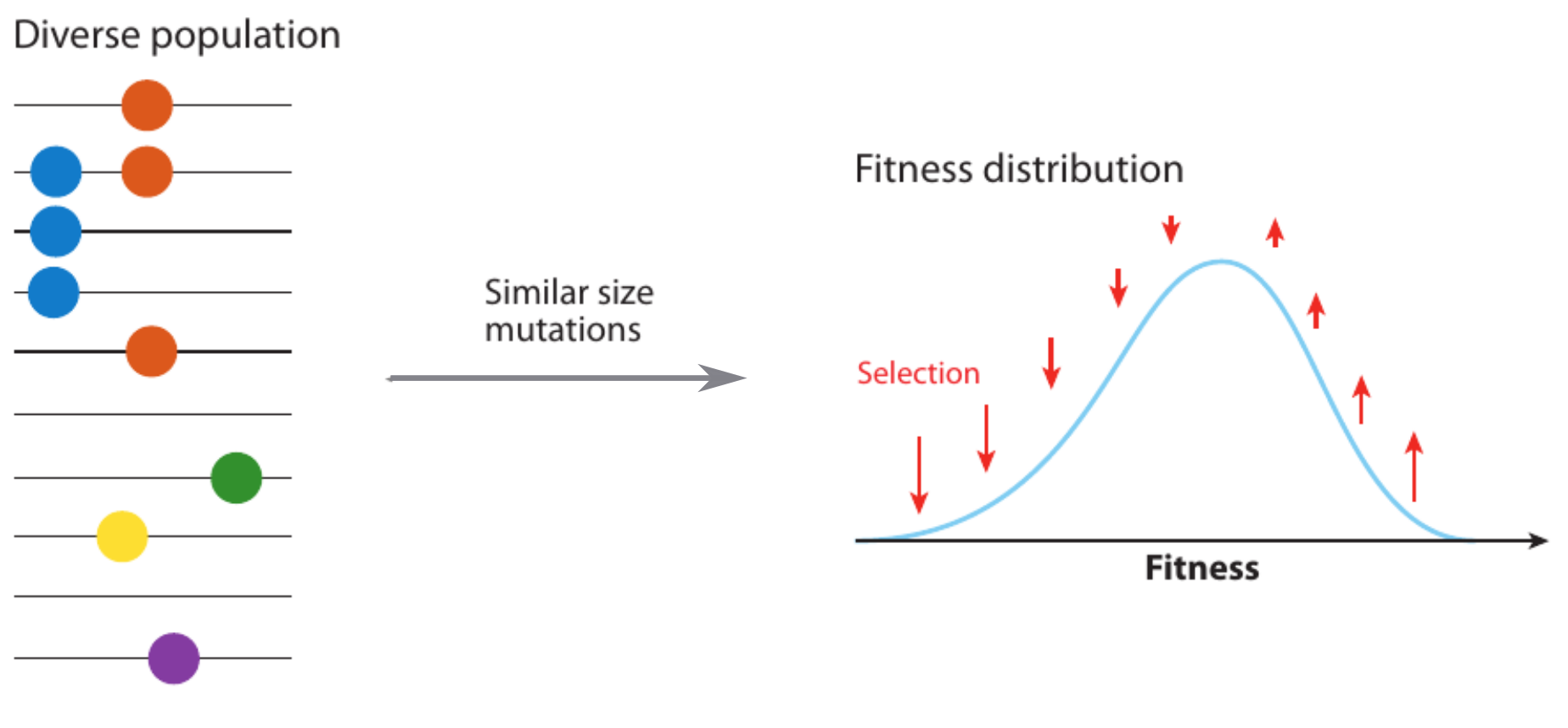
- Speed of adaptation is logarithmic in population size
- Environment (fitness landscape), not mutation supply, determines adaptation
- Different models have universal emerging properties
Neutral/Kingman coalescent
strong selection
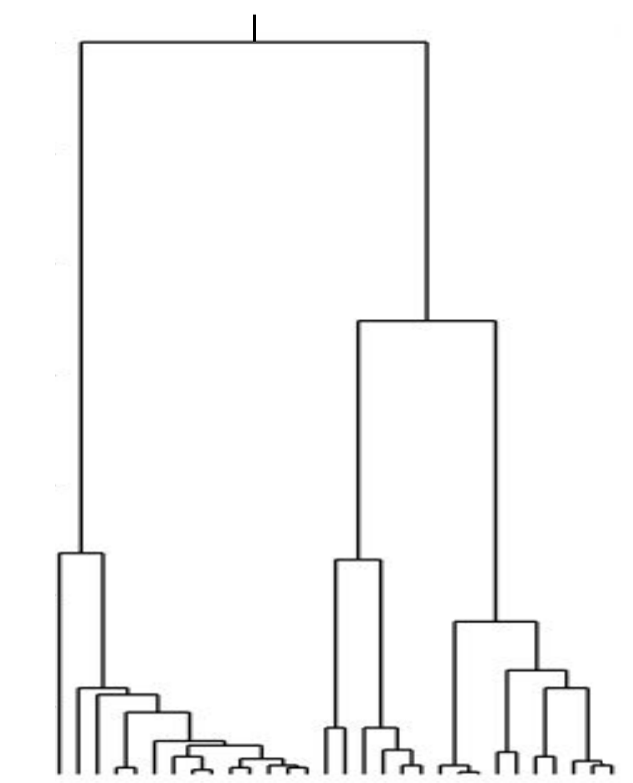

Bolthausen-Sznitman Coalescent
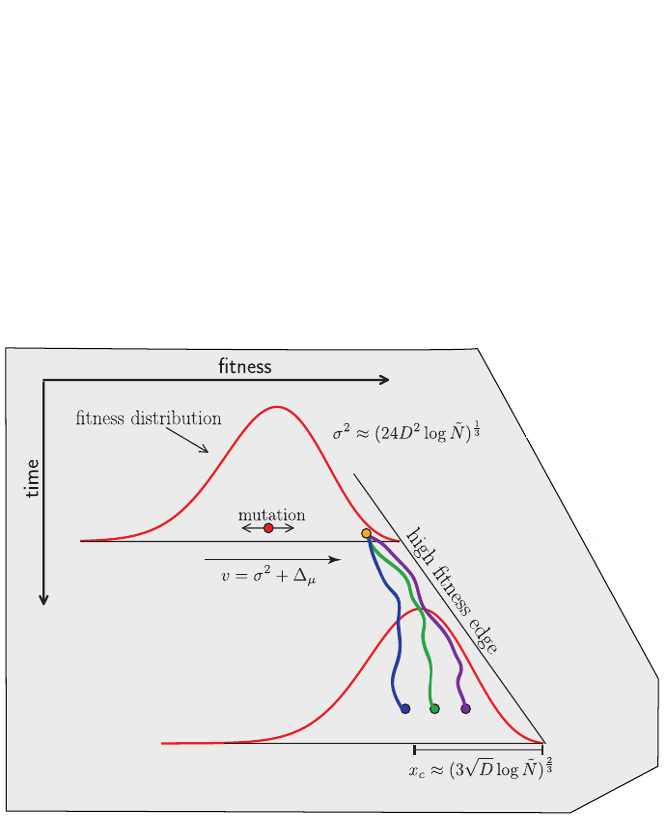
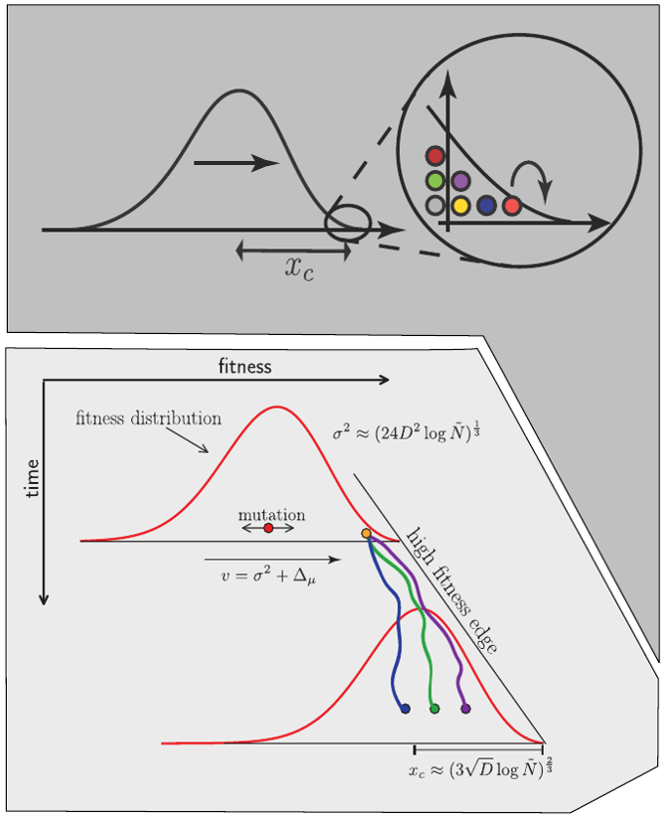
Burst in the tree ↔ high fitness
Predicting evolution

Given the branching pattern:
- can we predict fitness?
- pick the closest relative of the future?
Fitness inference from trees
$$P(\mathbf{x}|T) = \frac{1}{Z(T)} p_0(x_0) \prod_{i=0}^{n_{int}} g(x_{i_1}, t_{i_1}| x_i, t_i)g(x_{i_2}, t_{i_2}| x_i, t_i)$$
RN, Russell, Shraiman, eLife, 2014
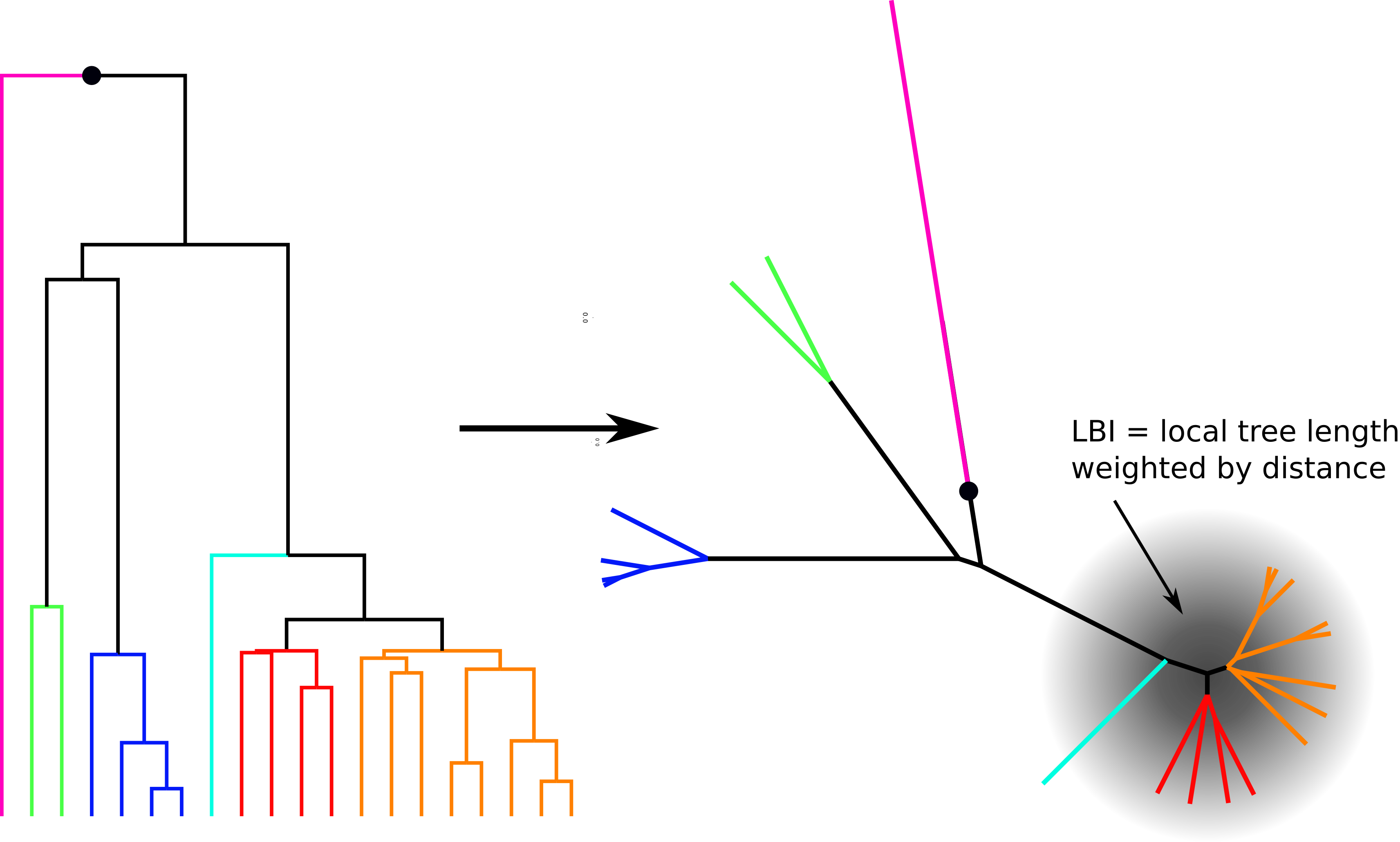
Simple heuristic: Local branching index
Validate on simulation data
- simulate evolution
- sample sequences
- reconstruct trees
- infer fitness
- predict ancestor of future
- compare to truth

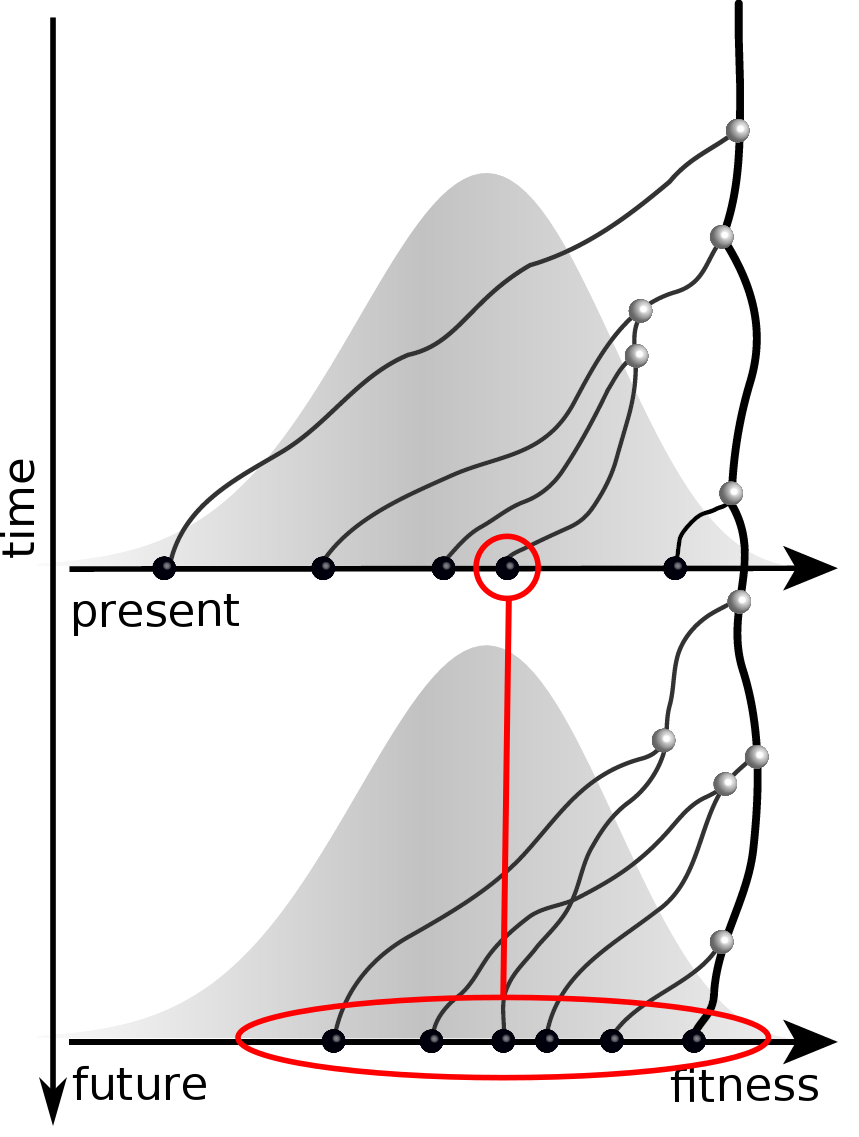
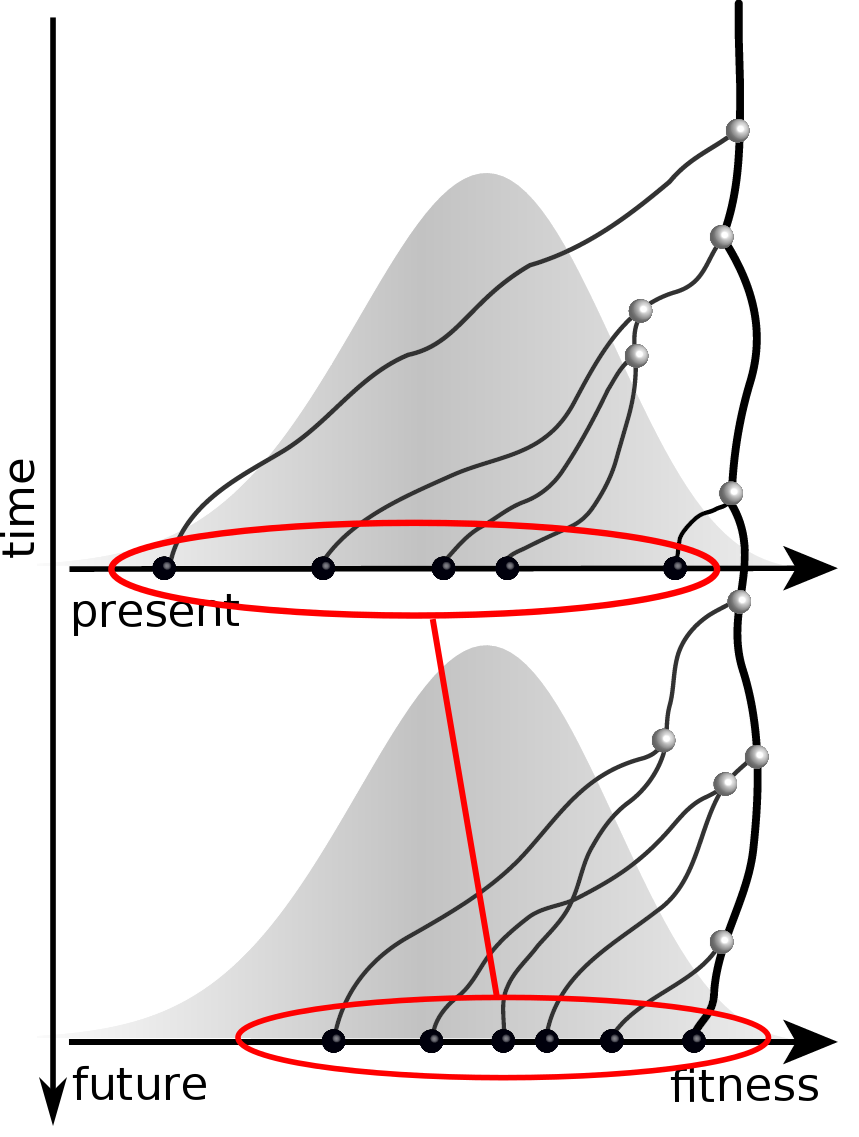
Validation on simulated data
Validation on simulated data
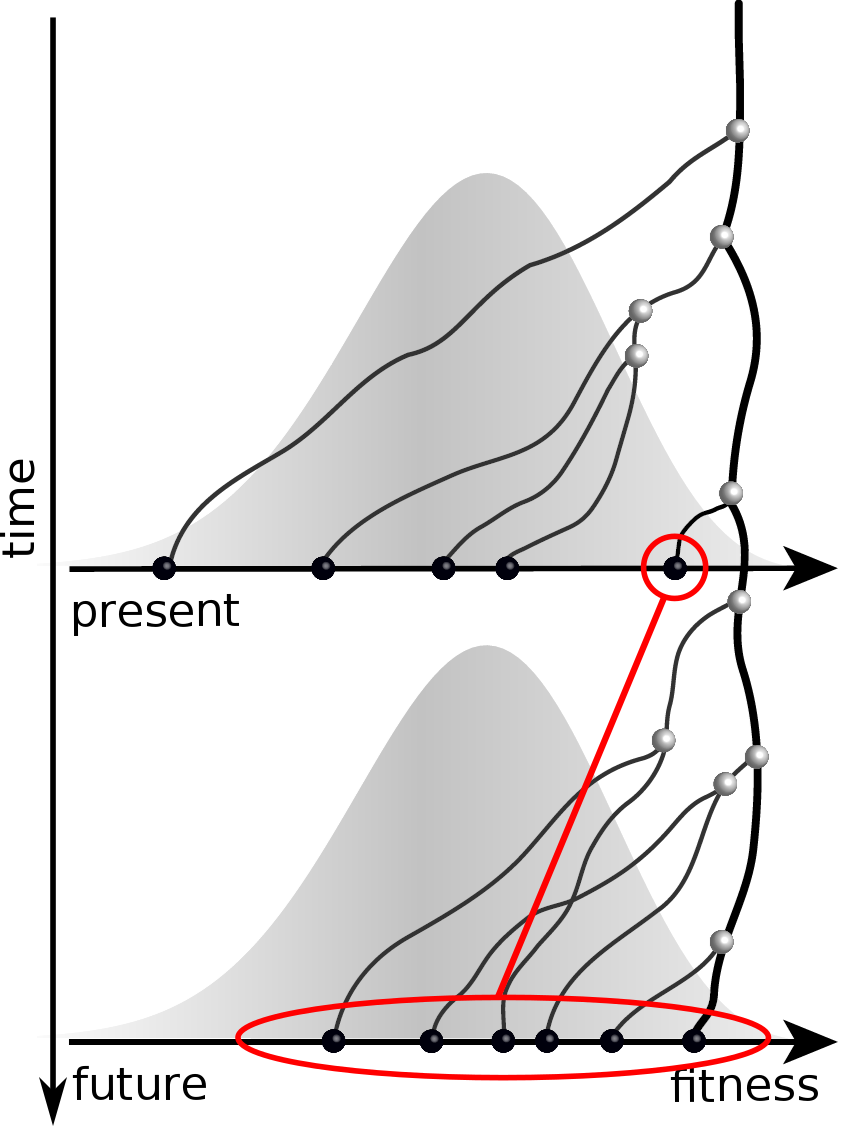
Prediction of the dominating H3N2 influenza strain
- no influenza specific input
- how can the model be improved? (see model by Luksza & Laessig)
- what other context might this apply?
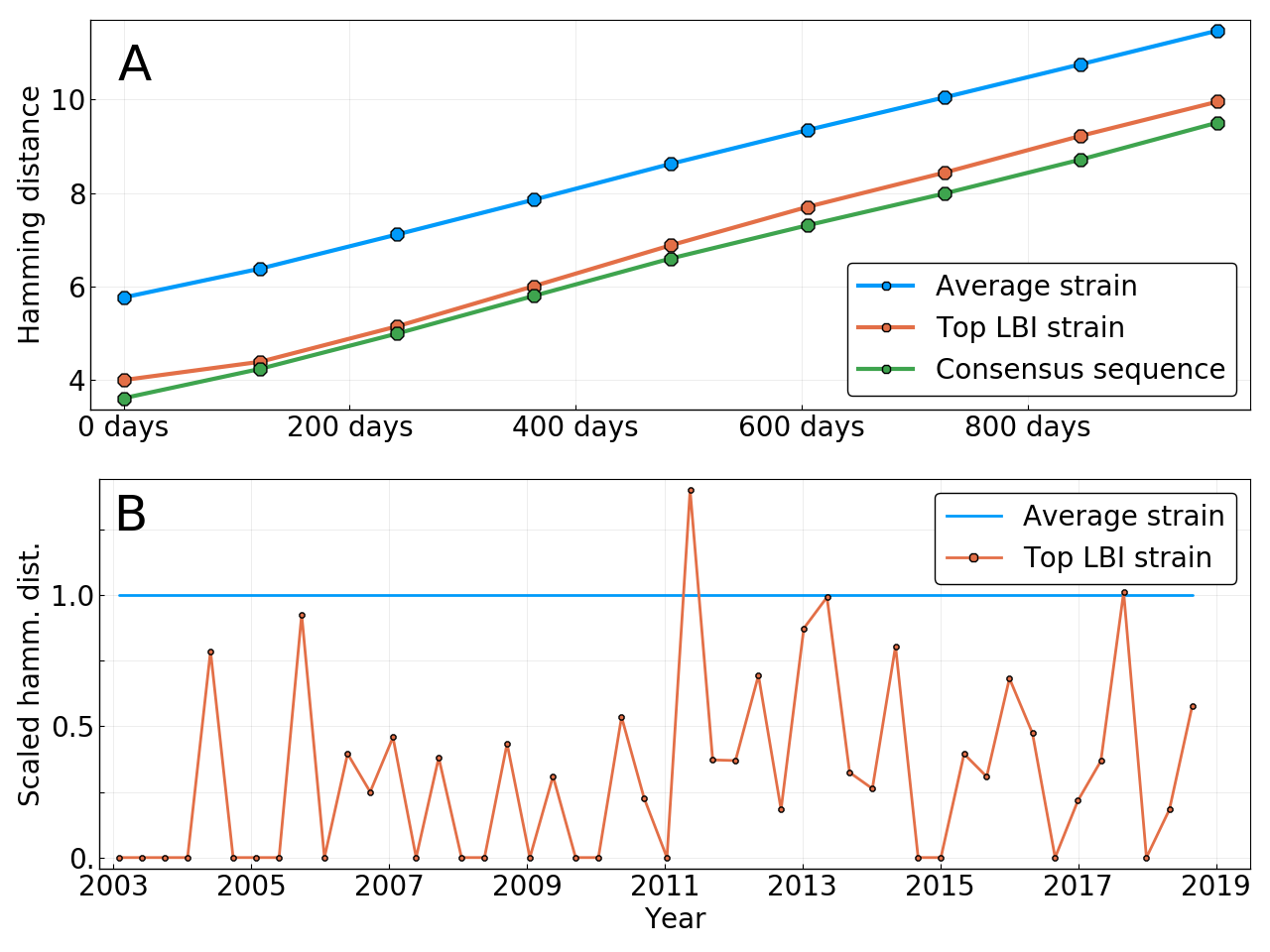
Limits of predictability
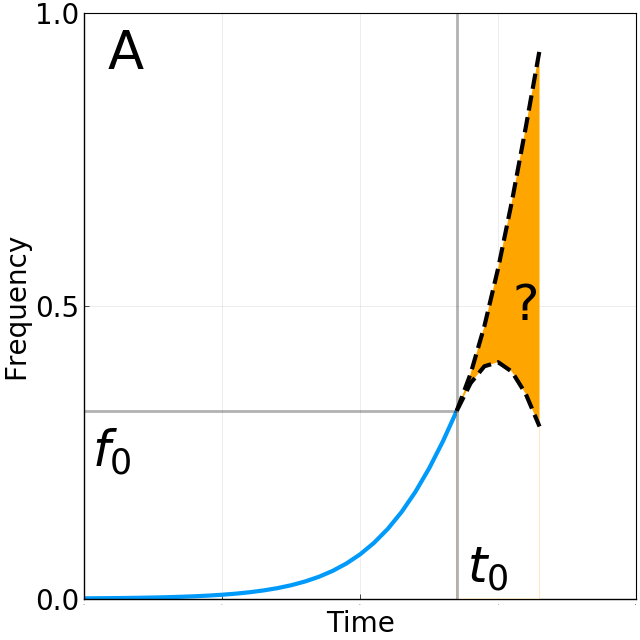
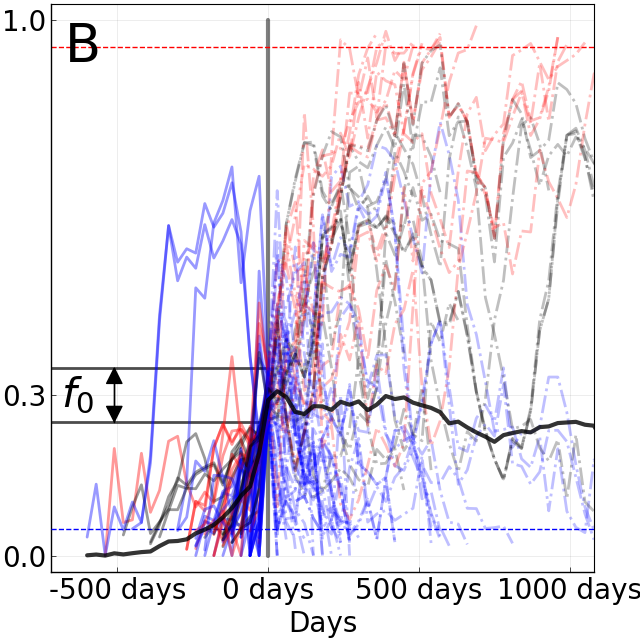
Limits of predictability
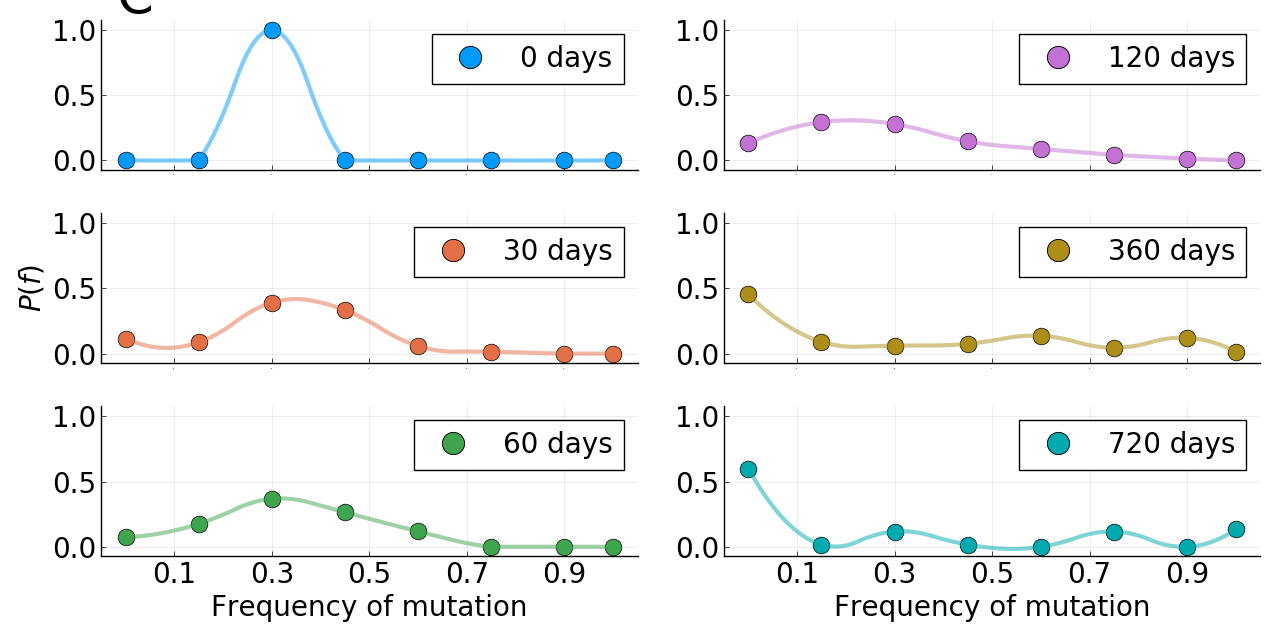
Limits of predictability
Influenza and Theory acknowledgments





- Boris Shraiman
- Colin Russell
- Trevor Bedford
- Pierre Barrat
- Oskar Hallatschek
- All the NICs and WHO CCs that provide influenza sequence data
- The WHO CCs in London and Atlanta for providing titer data


