SARS-CoV-2 und COVID19 -- genomische Epidemilogie in Echtzeit
Richard Neher
Biozentrum, University of Basel
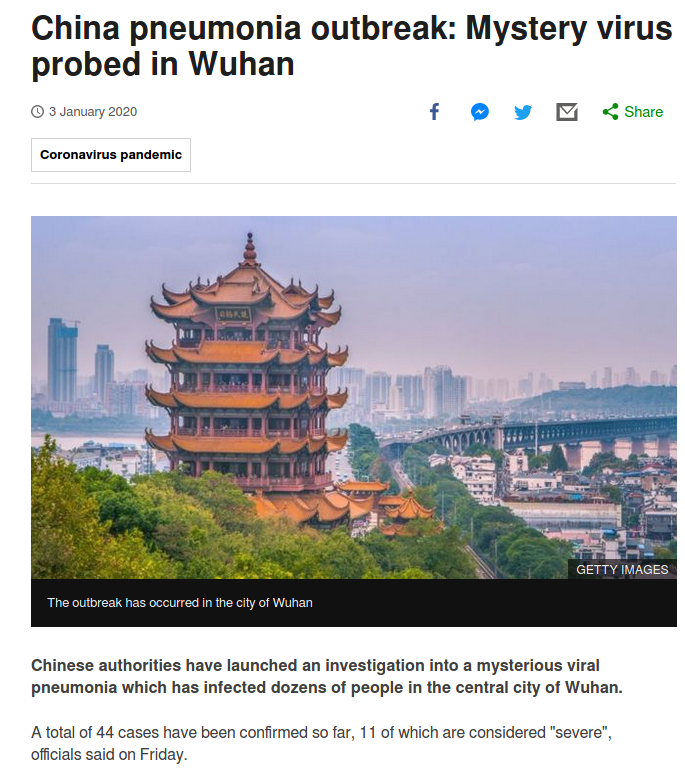 BBC
BBC
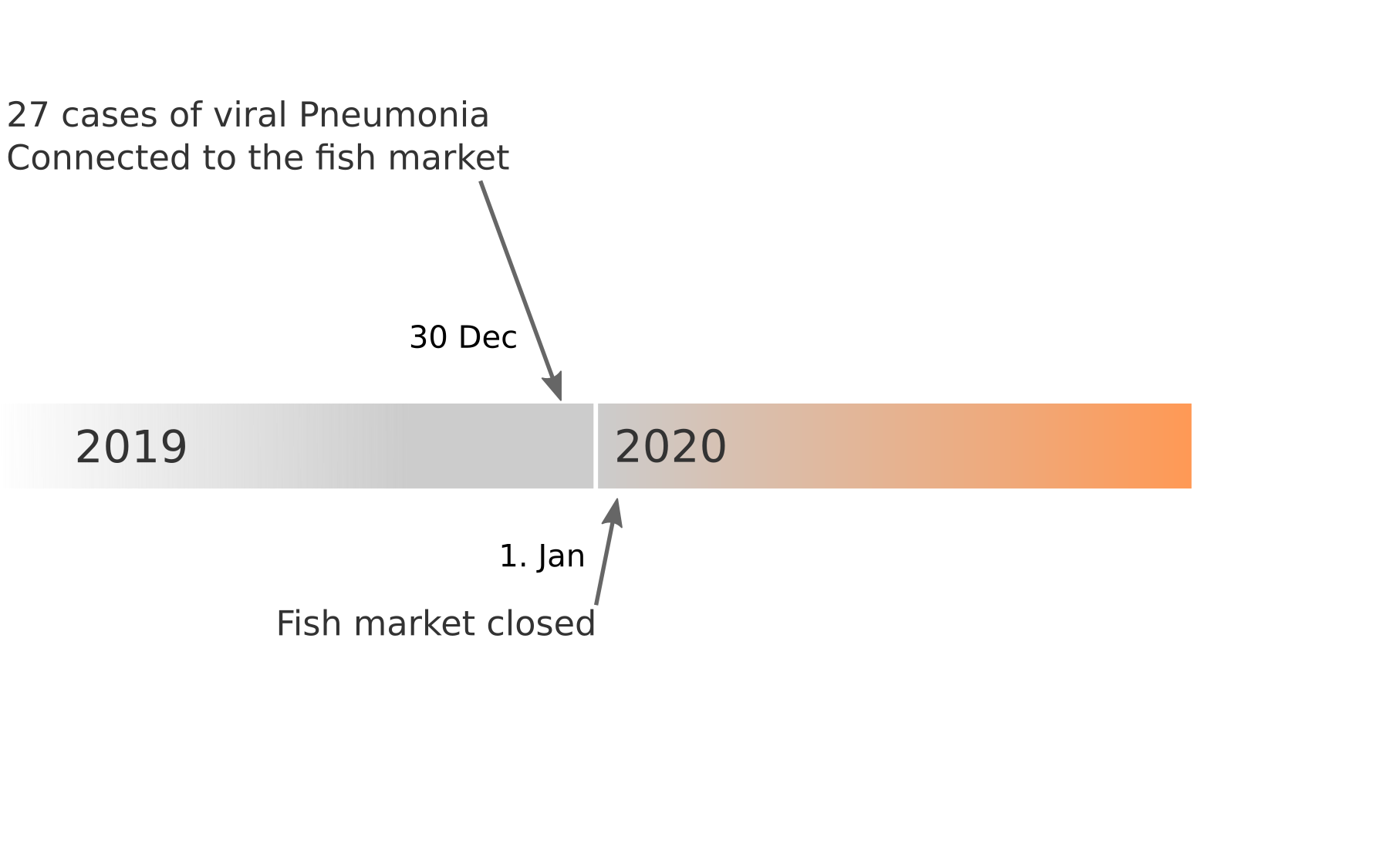 Data summarized by Ian MacKay
Data summarized by Ian MacKay
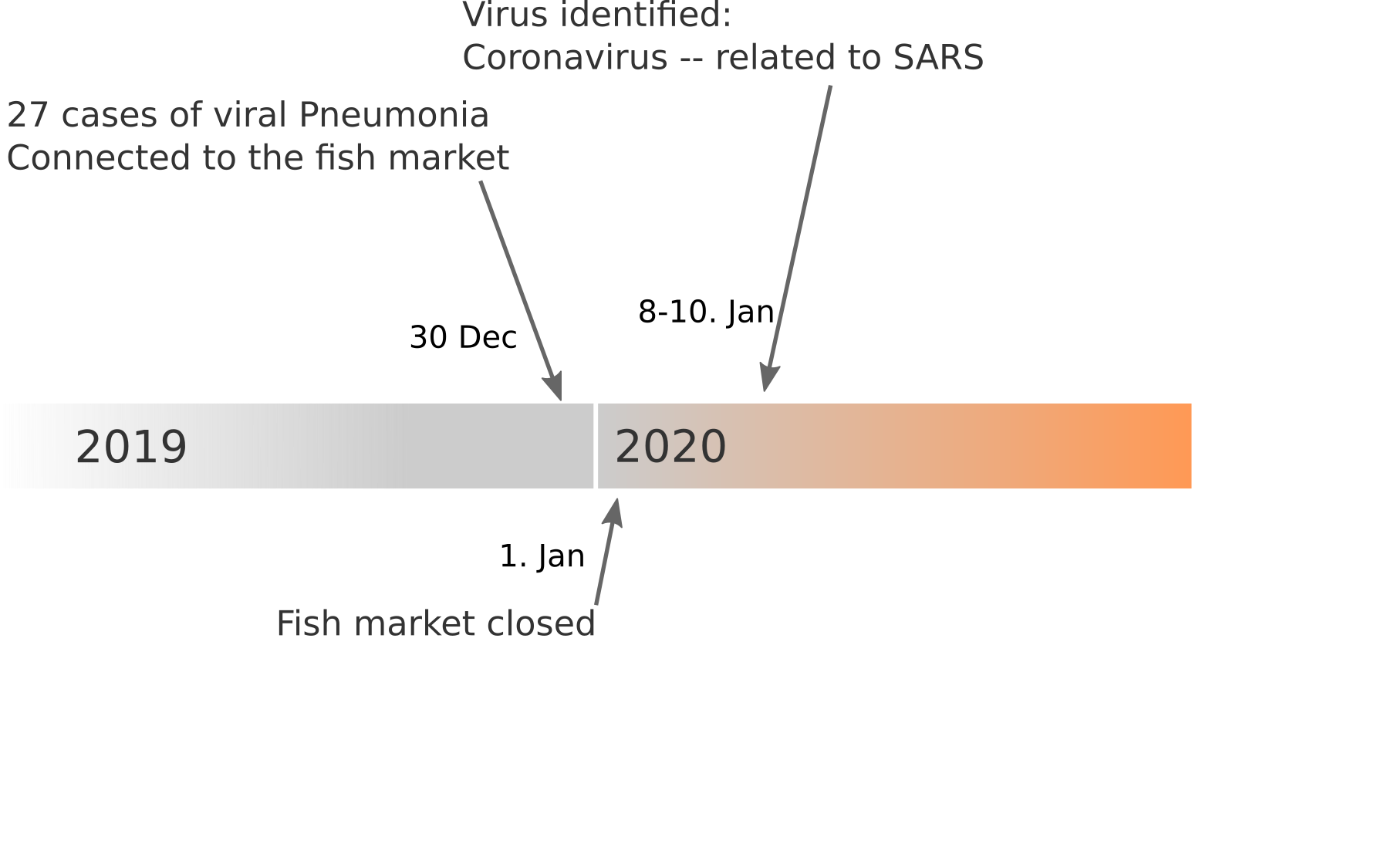 Data summarized by Ian MacKay
Data summarized by Ian MacKay
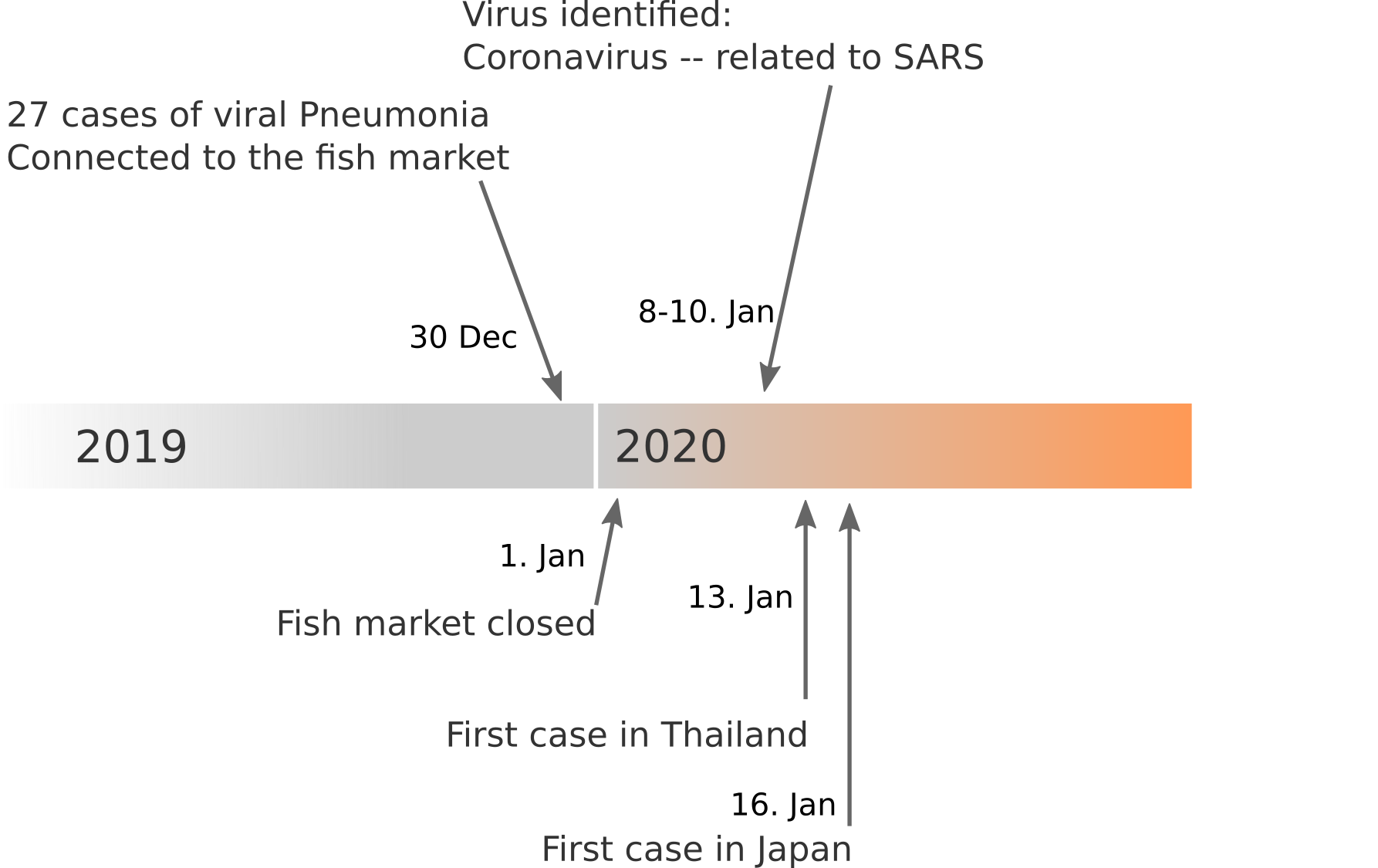 Data summarized by Ian MacKay
Data summarized by Ian MacKay
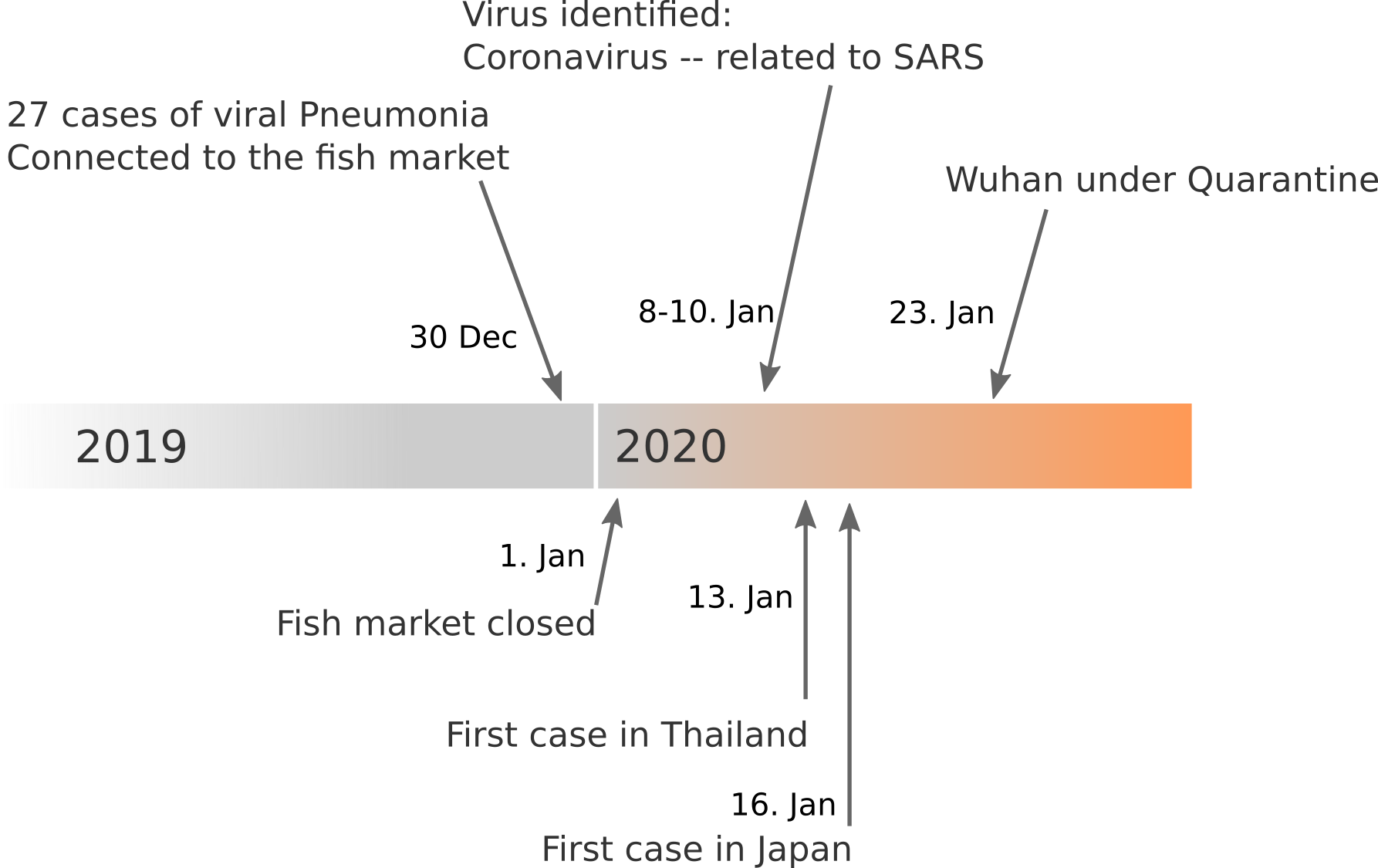 Data summarized by Ian MacKay
Data summarized by Ian MacKay
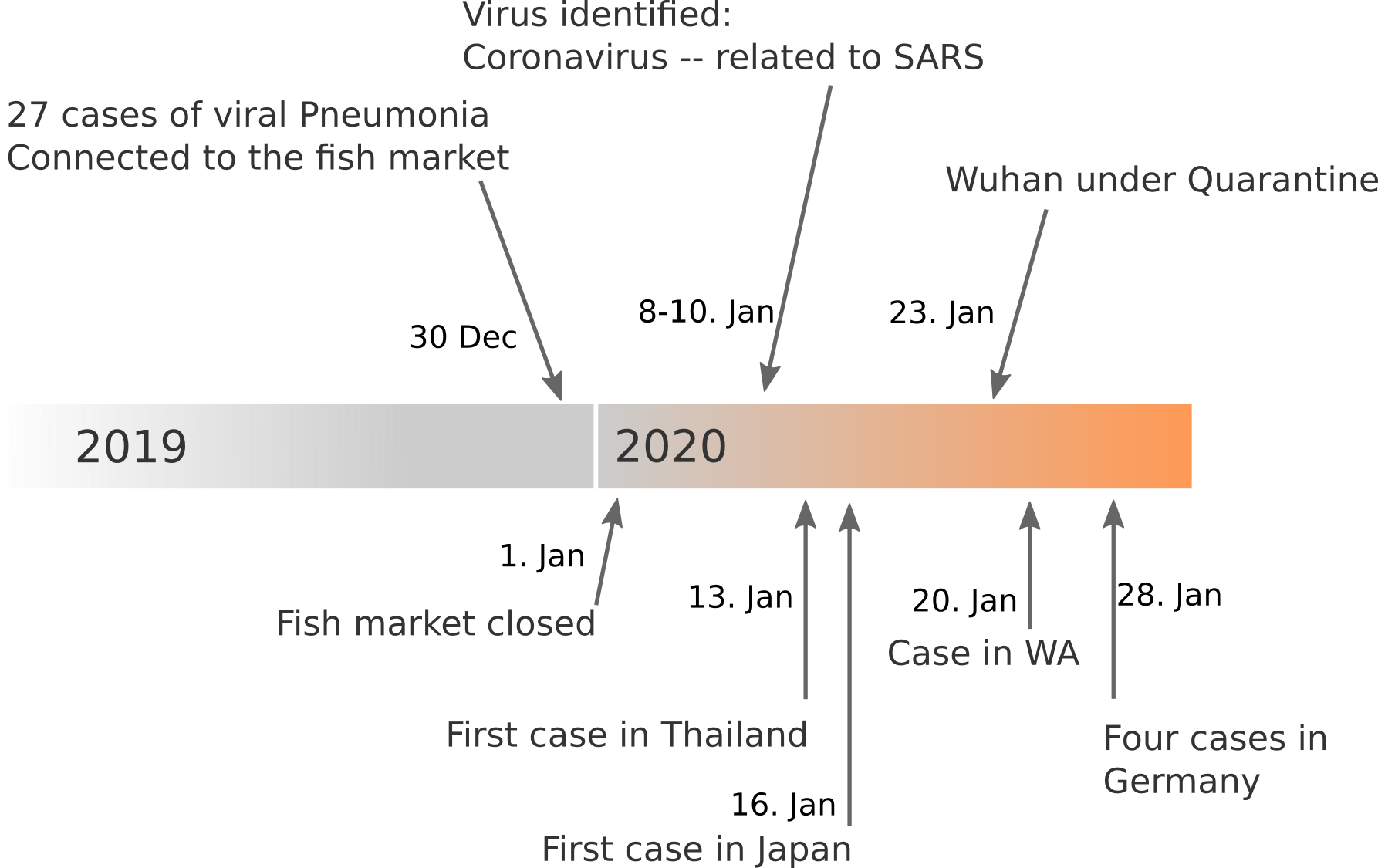 Data summarized by Ian MacKay
Data summarized by Ian MacKay
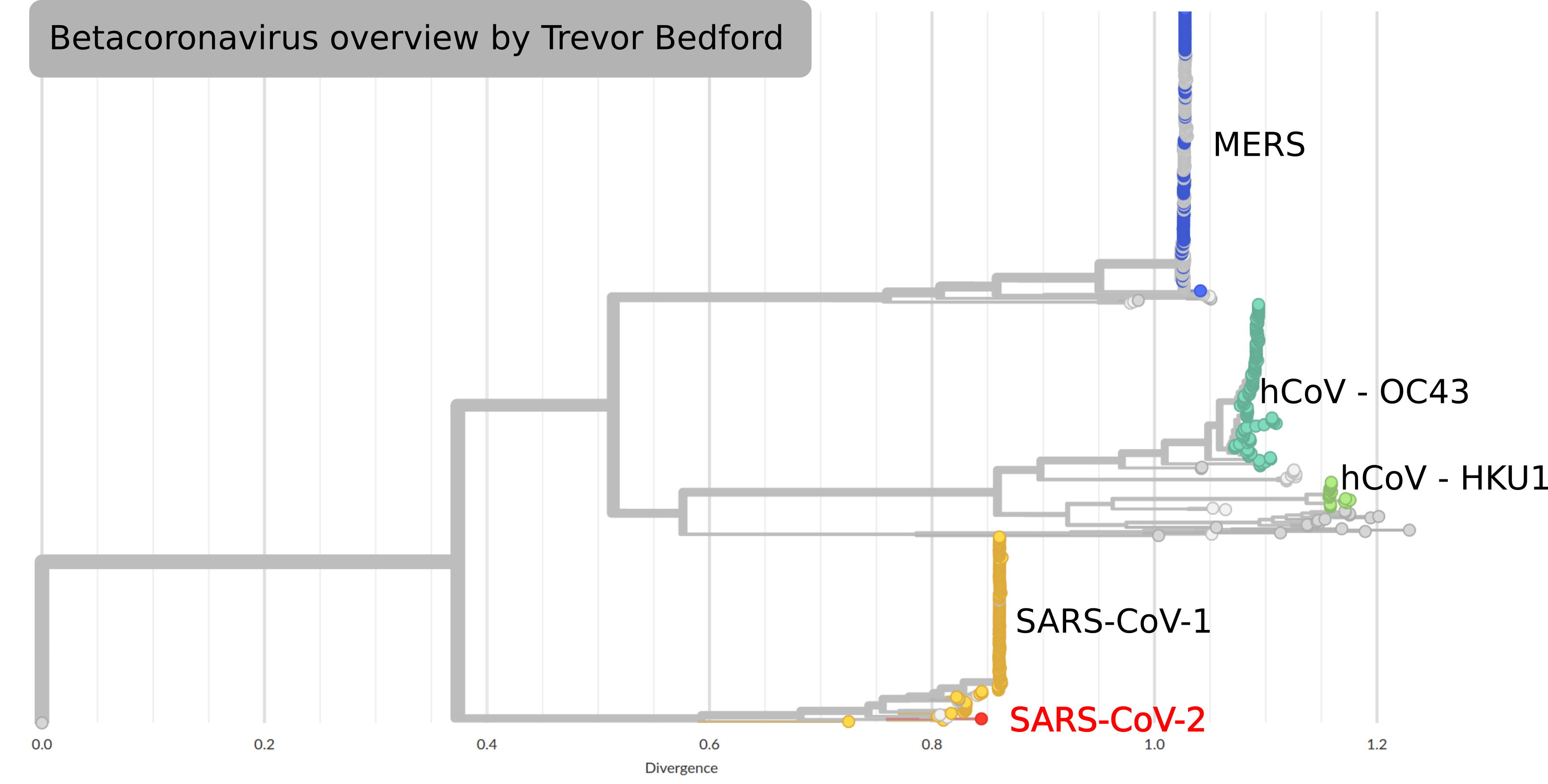 by Trevor Bedford
by Trevor Bedford
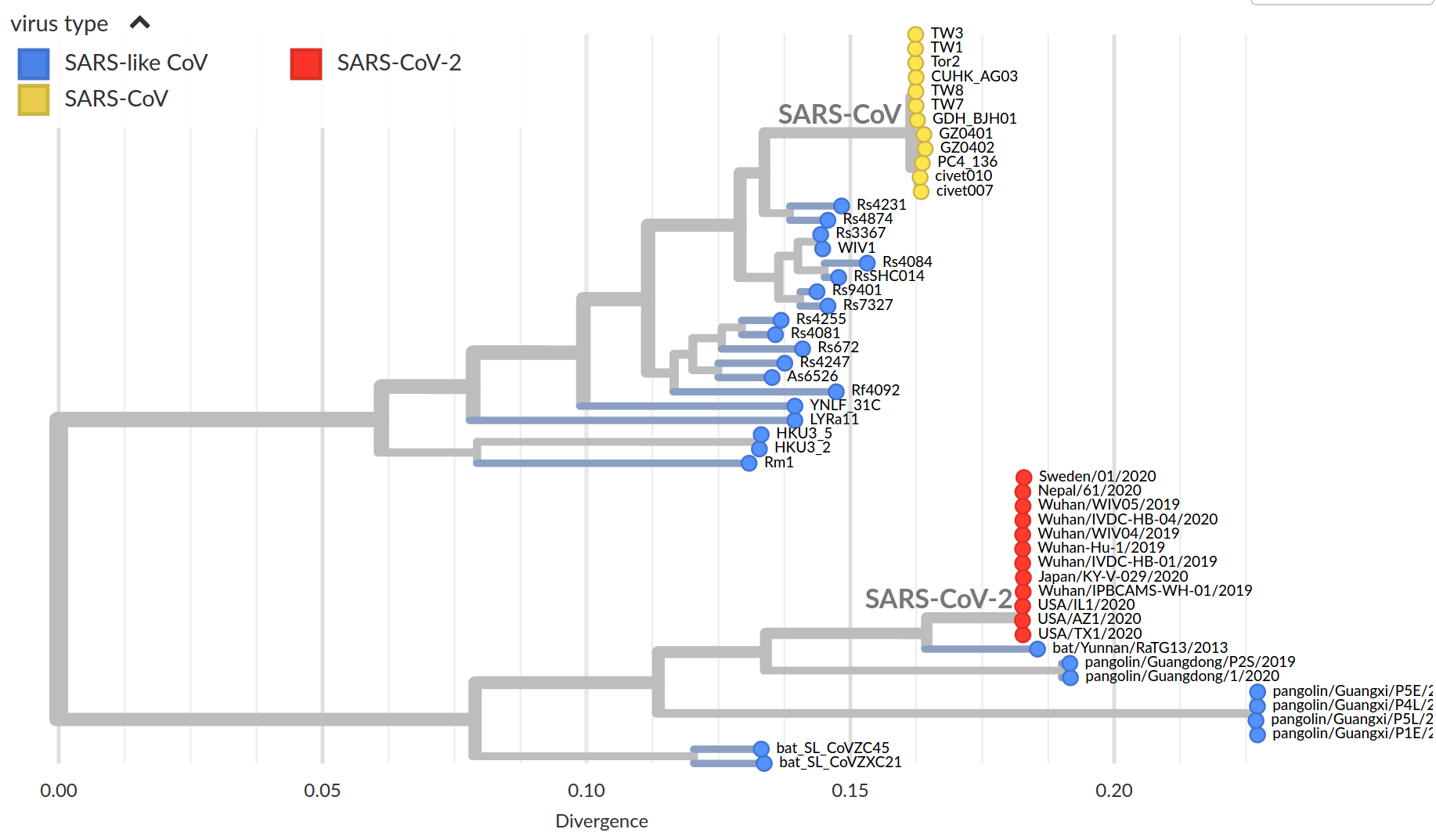 by Trevor Bedford
by Trevor Bedford
Tracking diversity and spread of SARS-CoV-2 in Nextstrain
Virus genomes change rapidly through time
A/Brisbane/100/2014
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
A/Brisbane/1000/2015
GGATAATTCTATTAACCATGAAGACTATTATTGCTTT...
A/Brisbane/1/2017
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
... hundreds of thousands of sequences...
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
A/Brisbane/1000/2015
GGATAATTCTATTAACCATGAAGACTATTATTGCTTT...
A/Brisbane/1/2017
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
... hundreds of thousands of sequences...
Genome sequences record the spread of pathogens




in SARS-CoV-2 a new mutation accumulates about every 2 weeks
Available data on Jan 26
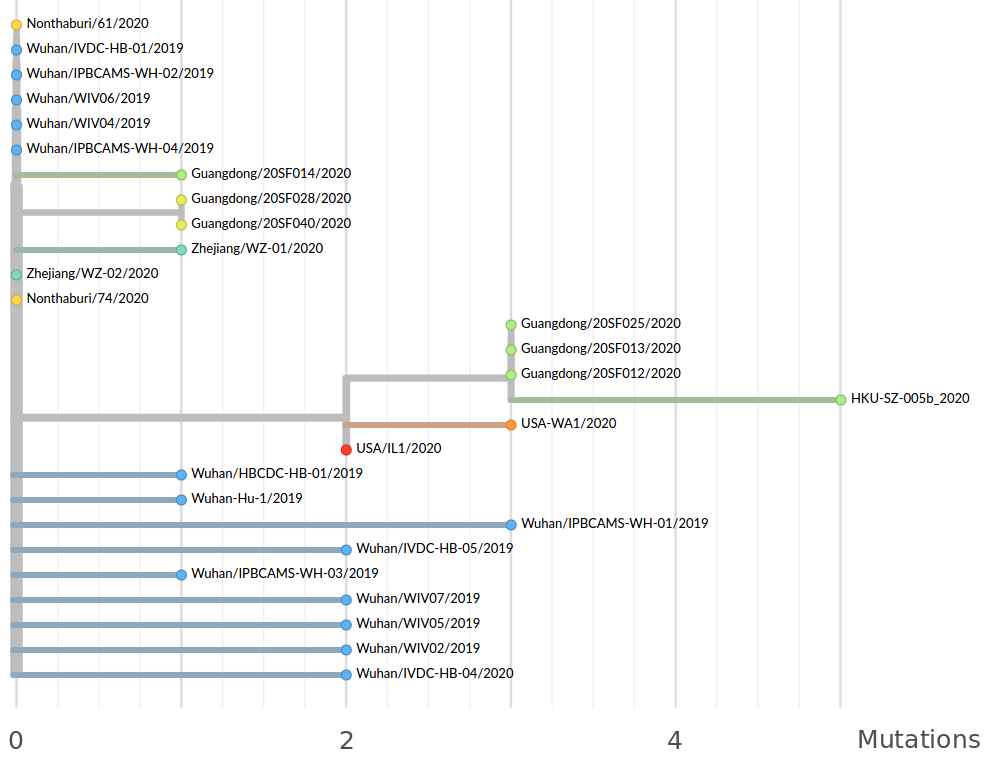
Early genomes differed by only a few mutations, suggesting very recent emergence
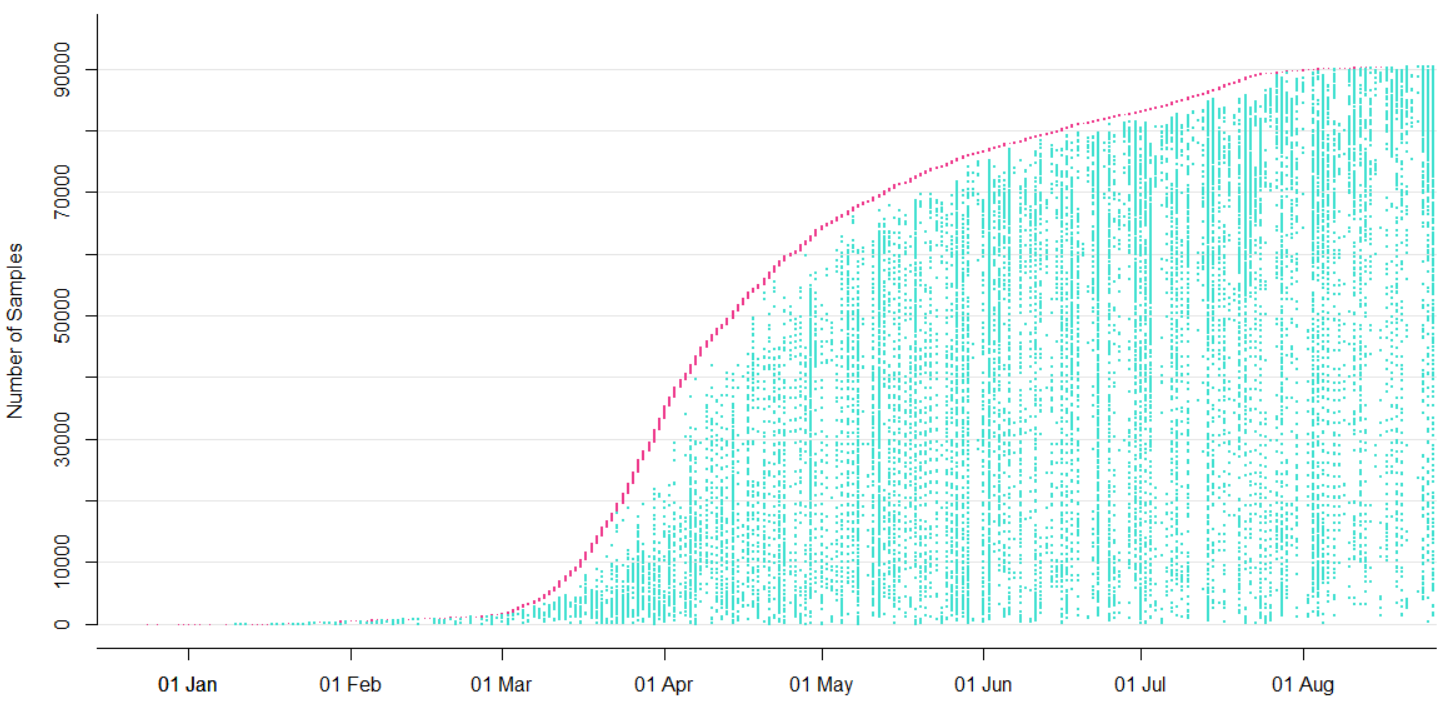
→ the closest to "real-time" we have experienced so far...
Figure by James Hadfield/Emma HodcroftTurning data into actionable insights
- hundreds of new sequences every day
- more than 200k sequences right now
- comprehensive analysis require hours to days to complete
→ requires continuous analysis and easy dissemination
→ interpretable and intuitive visualization
nextstrain.org
joint project with Trevor Bedford & his lab
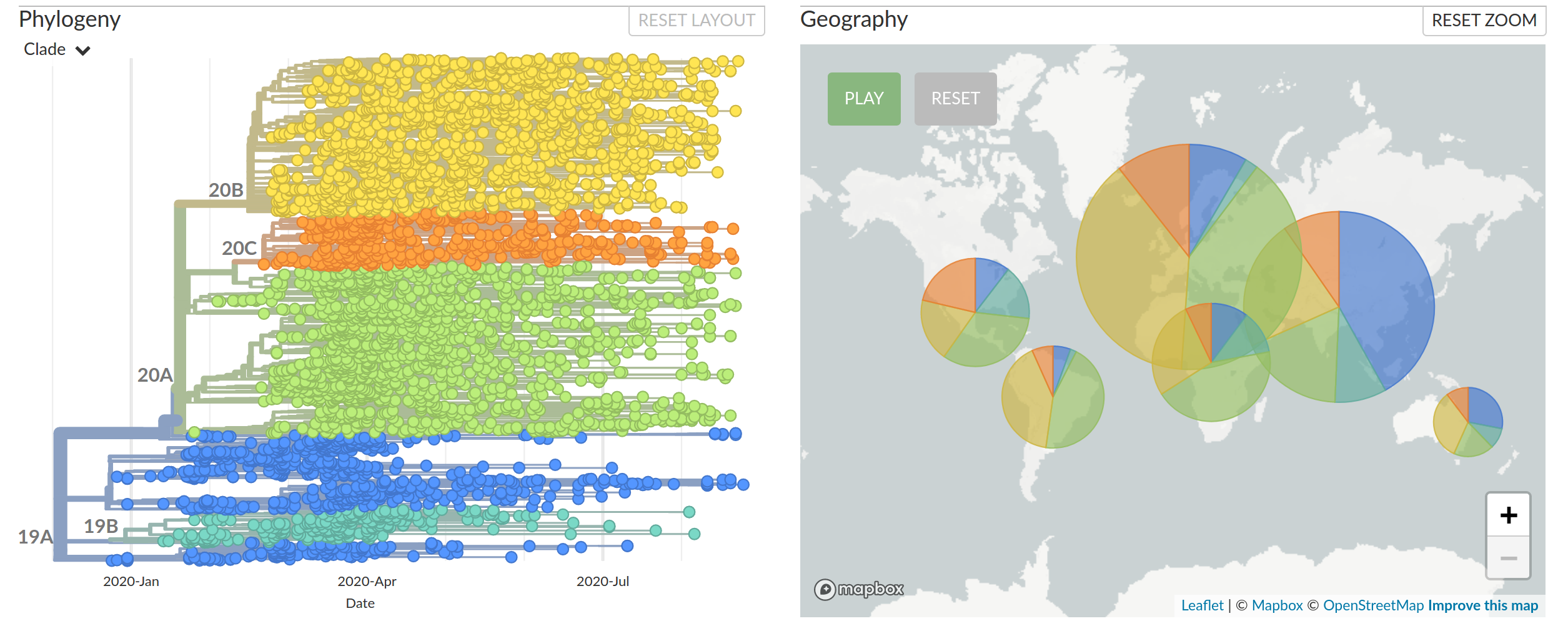
Diversified into multiple global variants. Groups 20A/B/C have taken over.
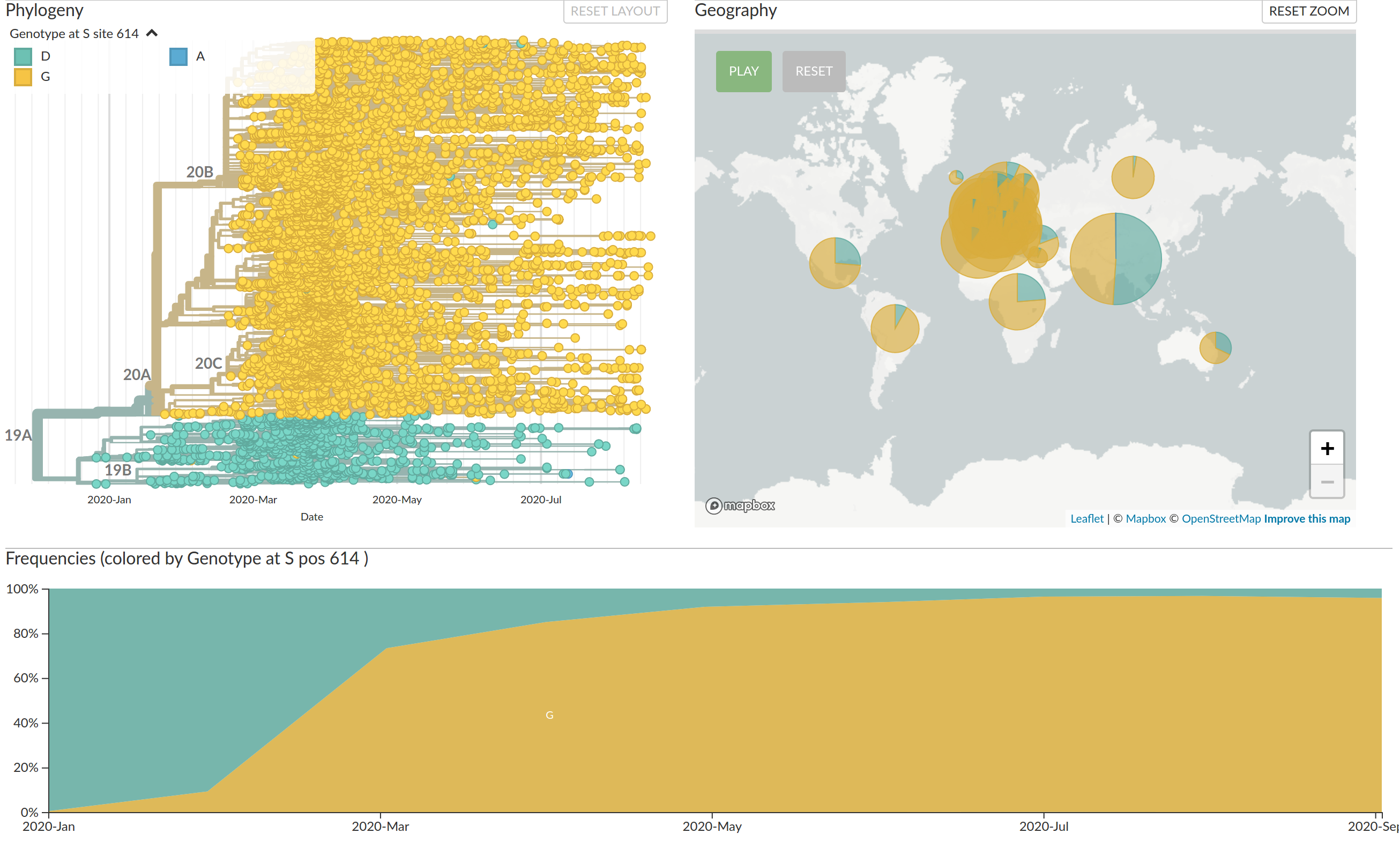
Large scale spread of the virus
Regional variants emerged over the summer
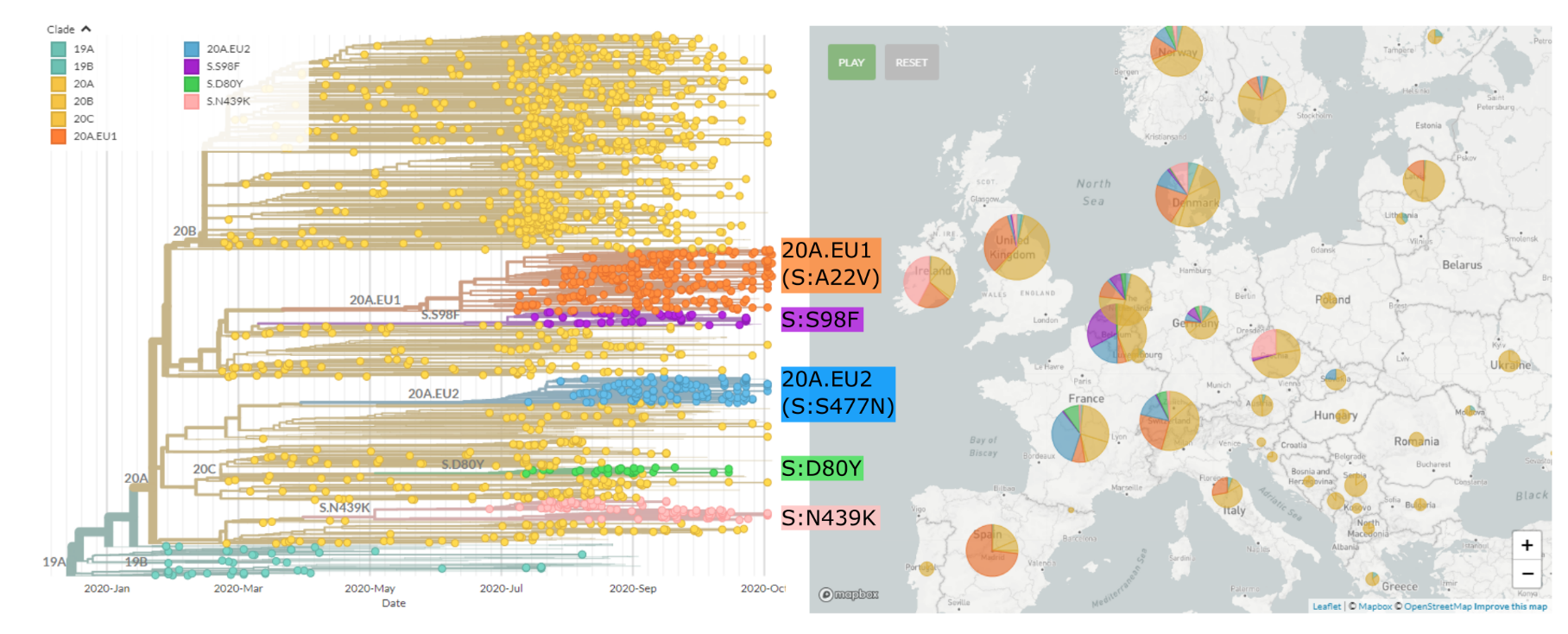
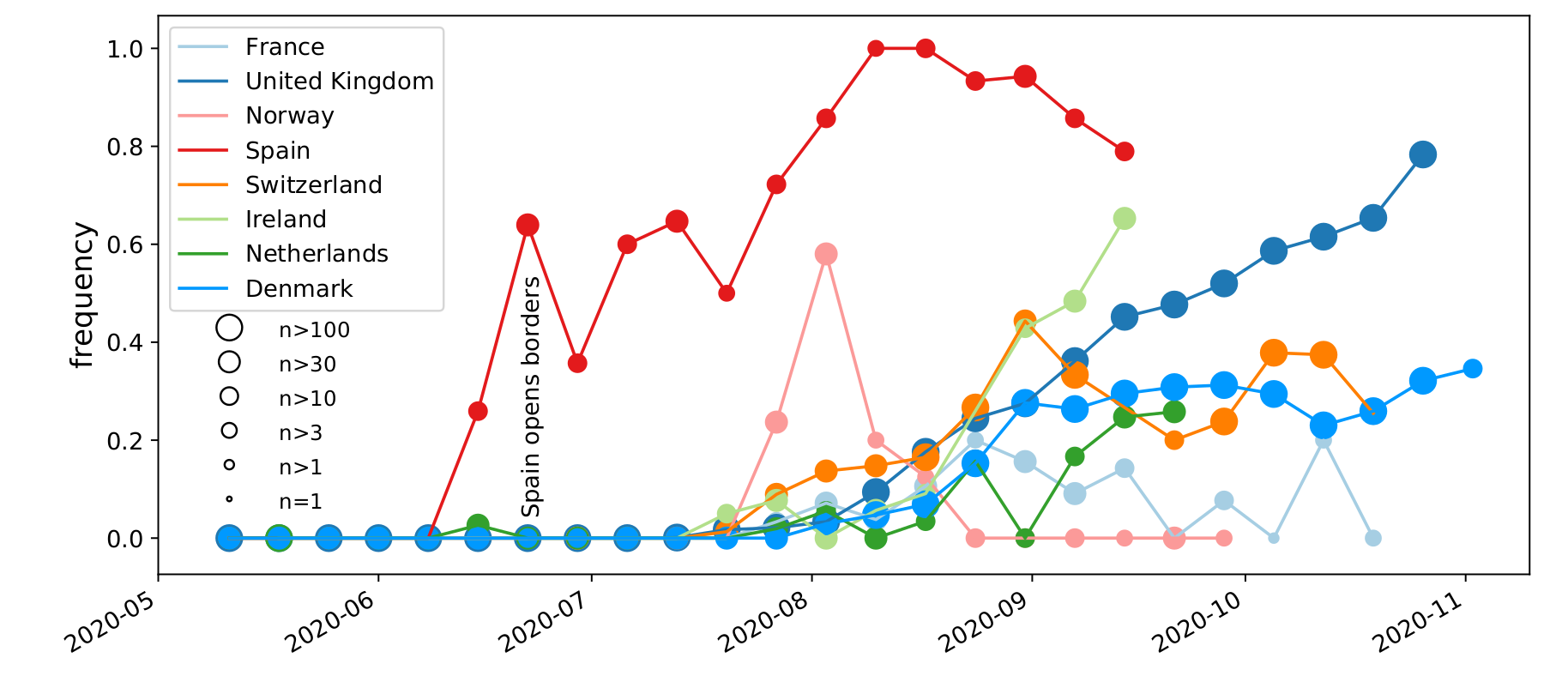
Spread of a variant through travel and tourism
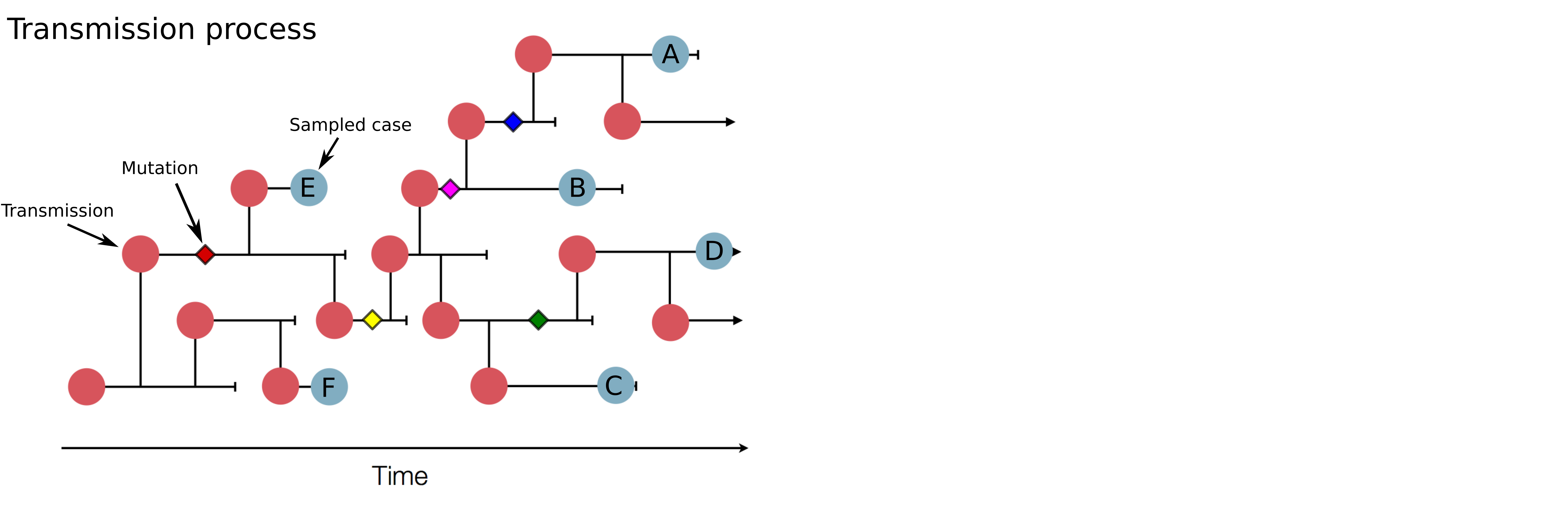
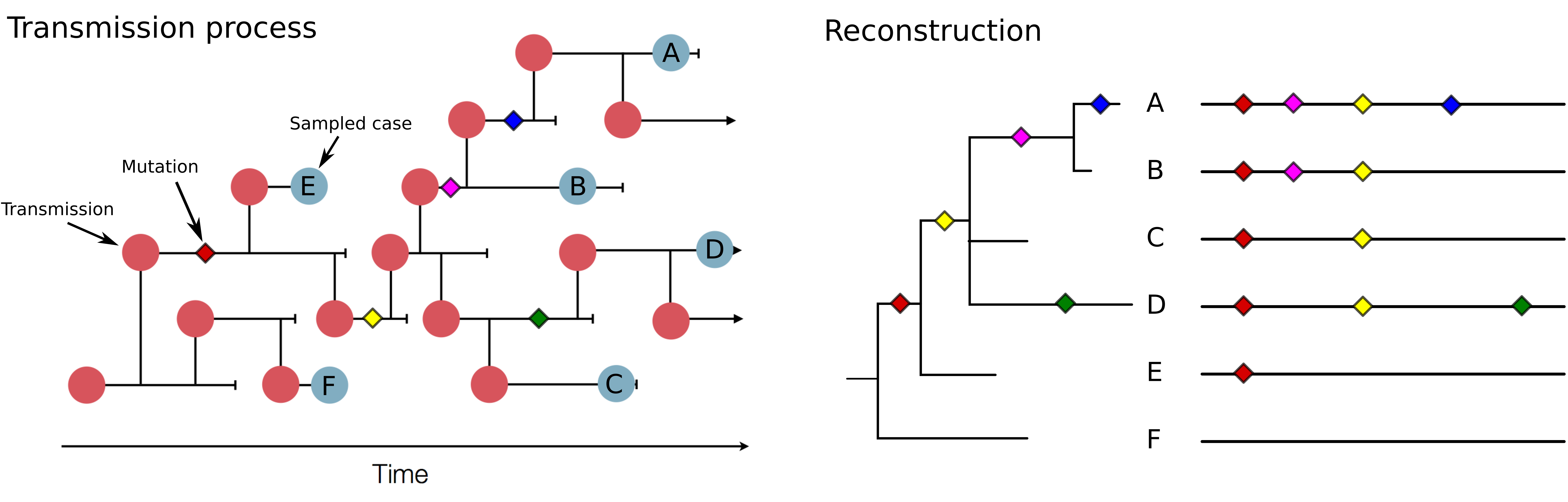
Genomic analysis as complement to contact tracing
Genomic analysis as complement to contact tracing
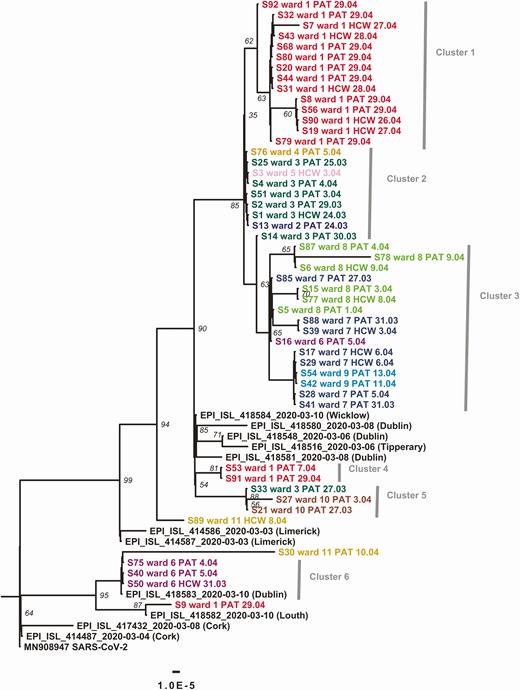
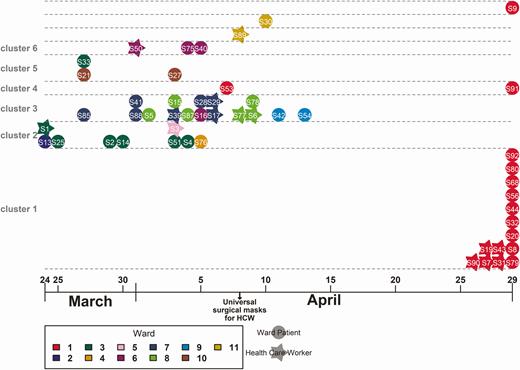
Investigation of hospital outbreaks
Influenza vaccine strain selection schedule
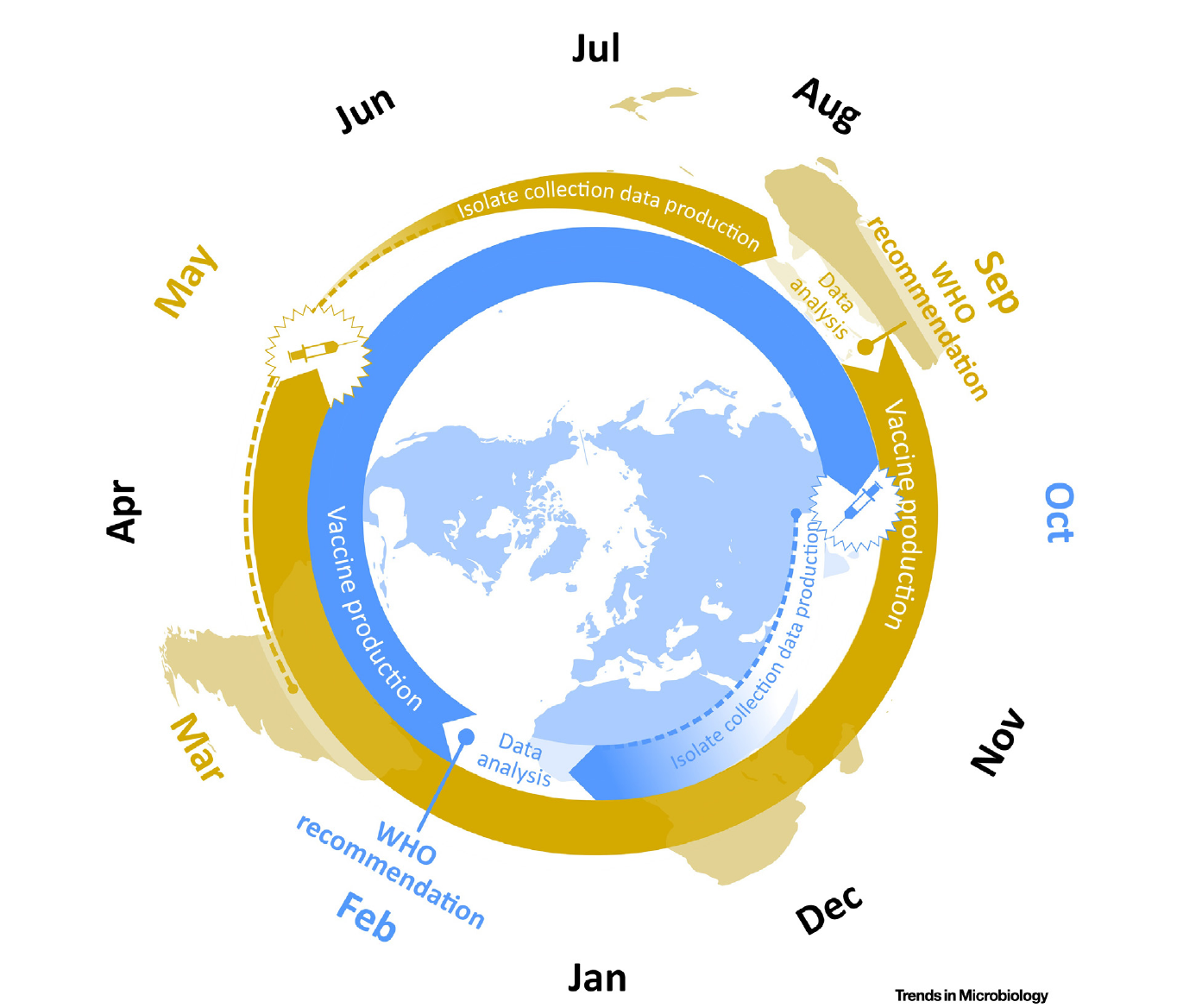 Klingen and McHardy, Trends in Microbiology
Klingen and McHardy, Trends in Microbiology
Herausforderungen und Zusammenfassung
- Offener Datenaustausch und Urheberschaft stehen immer wieder im Konflikt
- Analysen sind nicht einfach zu interpretieren
- Grosse Datenmengen, anspruchsvolle Methoden
- Zeitnahe und automatisierte Analysen sind essentiell
- Dennoch: Wir sollten Möglichkeiten, nicht Herausfordungen, sehen!
- Integration in die Routine-Surveillance
Acknowledgments
Trevor Bedford and his lab -- terrific collaboration since 2014

especially James Hadfield, Emma Hodcroft, Ivan Aksamentov, Moira Zuber, and Tom Sibley
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID


Acknowledgments
- Emma Hodcroft
- Moira Zuber
- Inaki Comas
- Fernando Gonzales
- Tanja Stadler
- Sarah Nadeau
- Tim Vaughan
- Jesse Bloom
- David Veesler
