Population genetics of rapid adaptation and influenza virus evolution
Richard Neher
Biozentrum & SIB, University of Basel
slides at neherlab.org/202101_.html
Human seasonal influenza viruses




- Influenza viruses evolve to avoid human immunity
- Vaccines need frequent updates

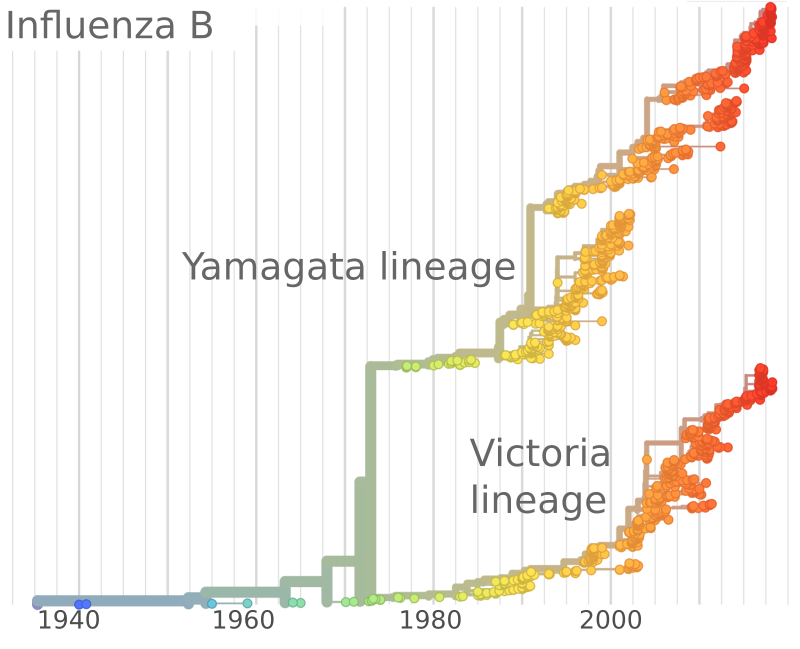
Influenza B viruses have split into two lineages
Fitness variation in rapidly adapting populations
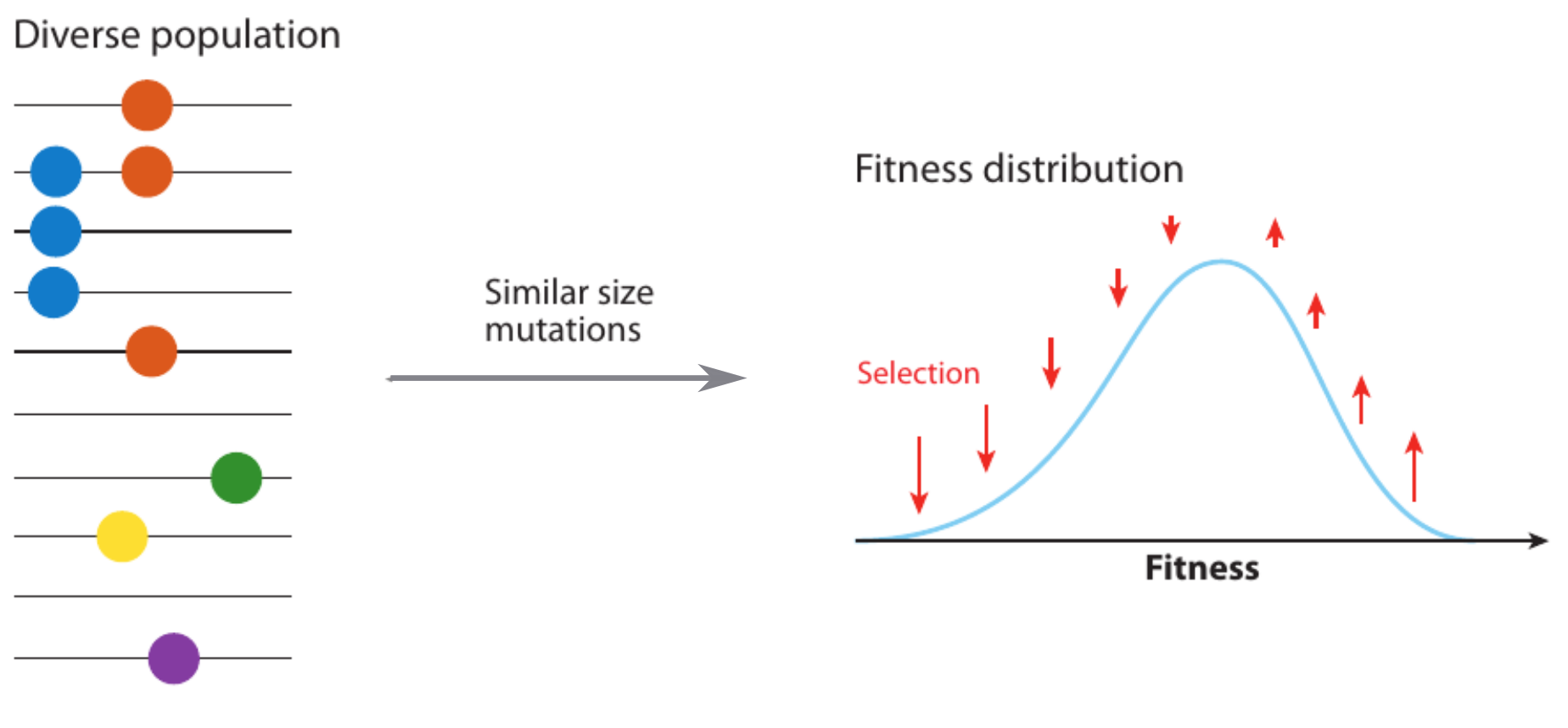
- Speed of adaptation is logarithmic in population size
- Environment (fitness landscape), not mutation supply, determines adaptation
- Different models have universal emerging properties
Neutral/Kingman coalescent
strong selection
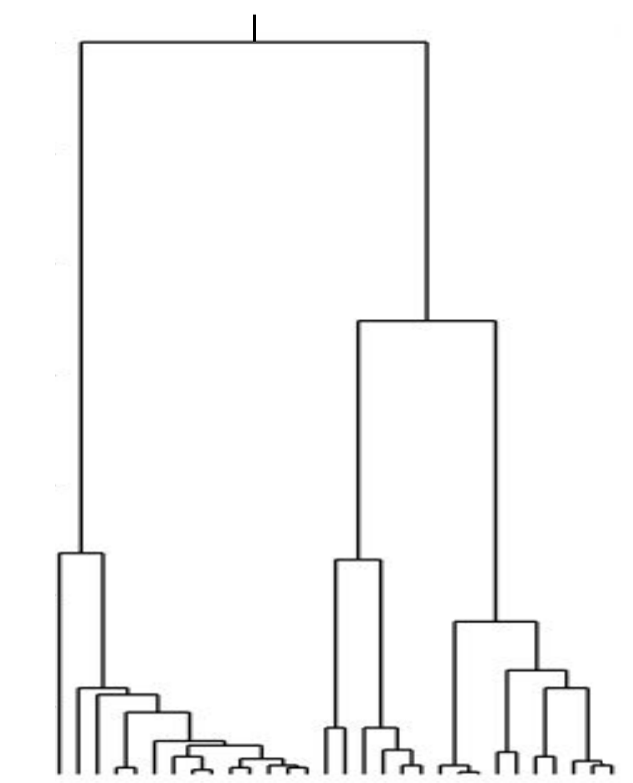

Bolthausen-Sznitman Coalescent
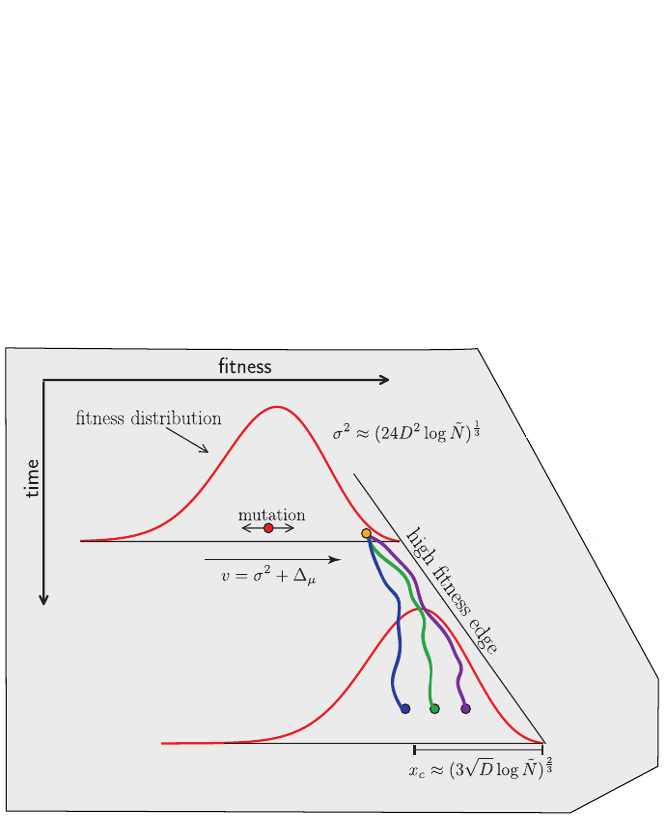
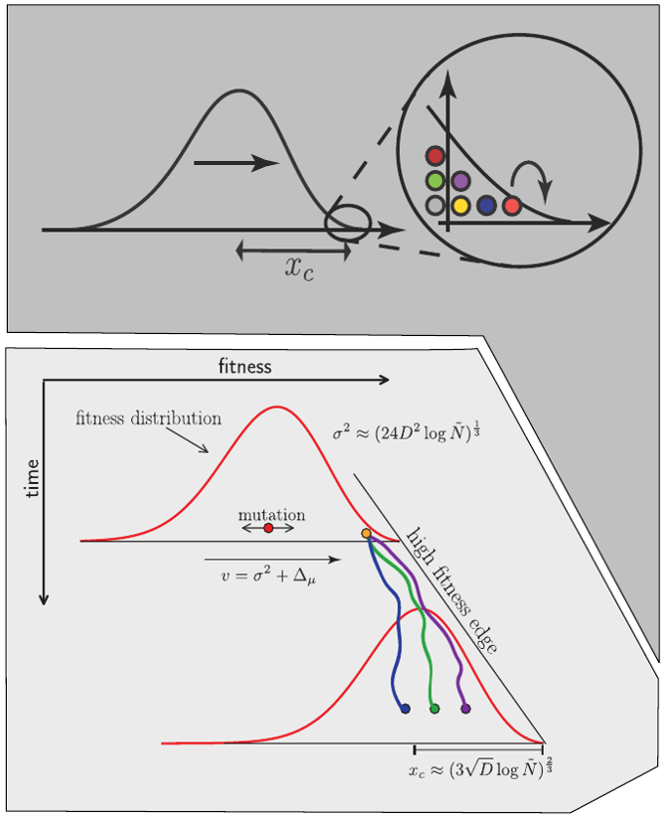
Burst in the tree ↔ high fitness
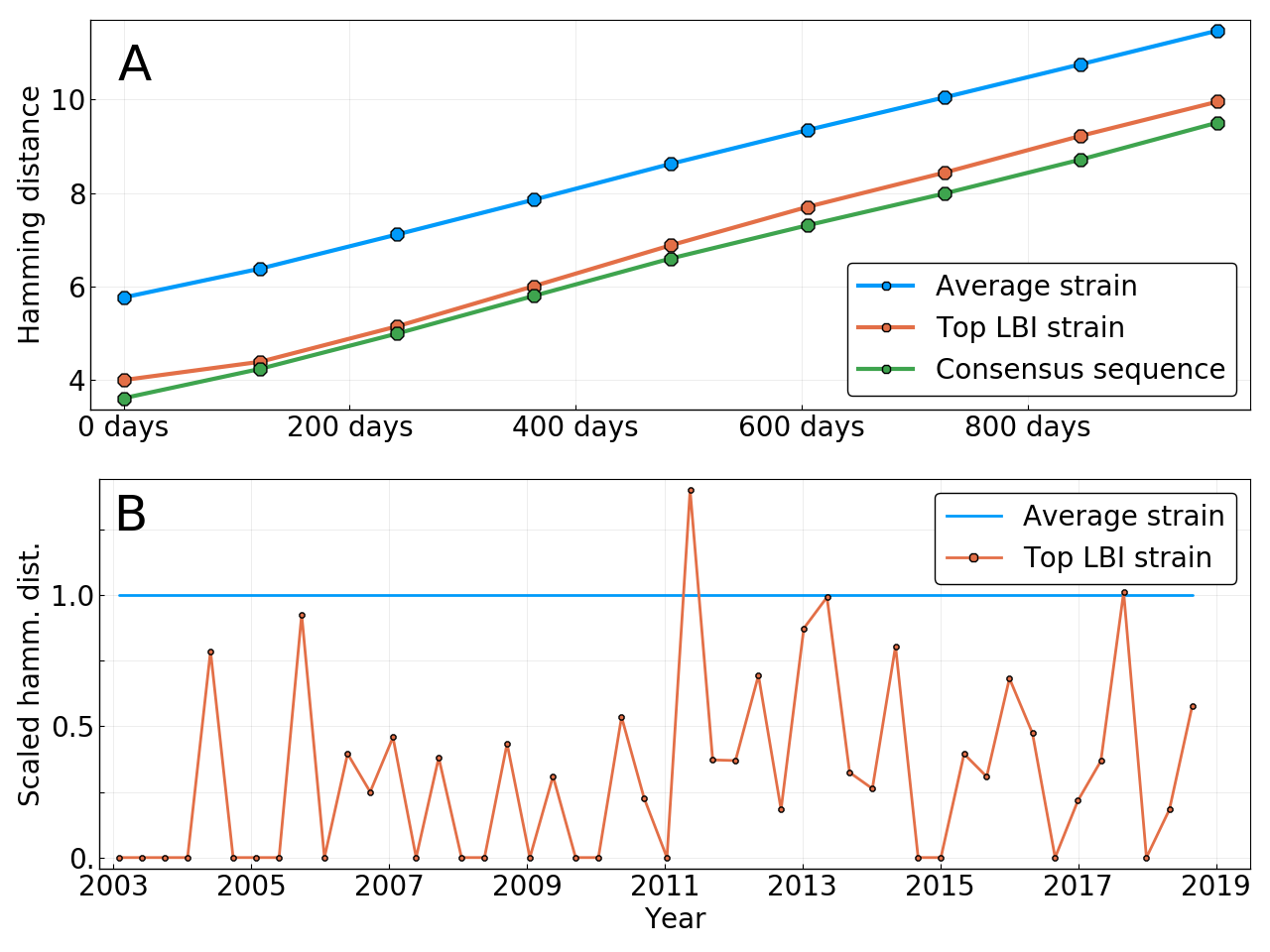
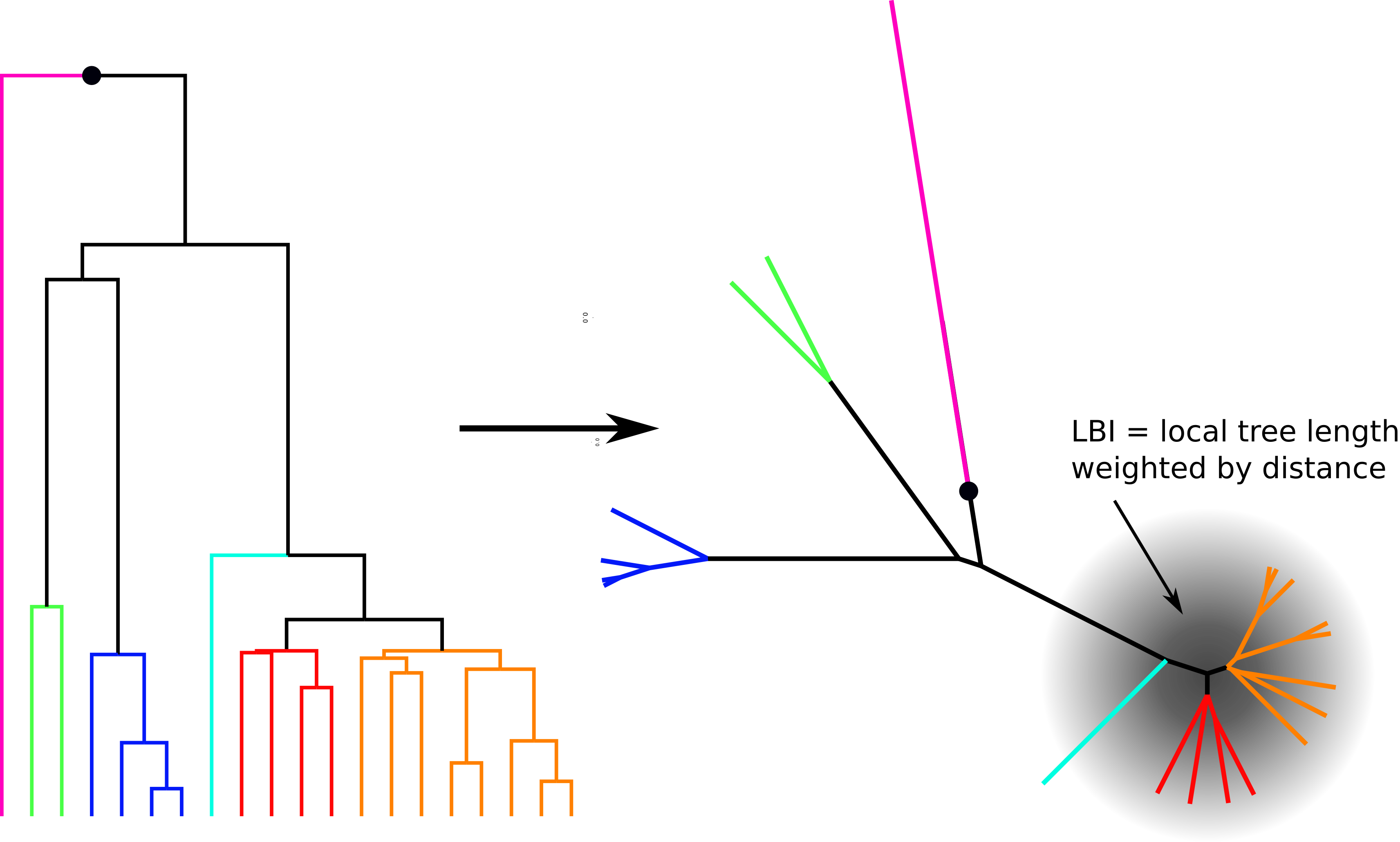
Simple heuristic: Local branching index
Prediction of the dominating H3N2 influenza strain
- no influenza specific input
- how can the model be improved? (see model by Luksza & Laessig)
- what other context might this apply?
Limits of predictability
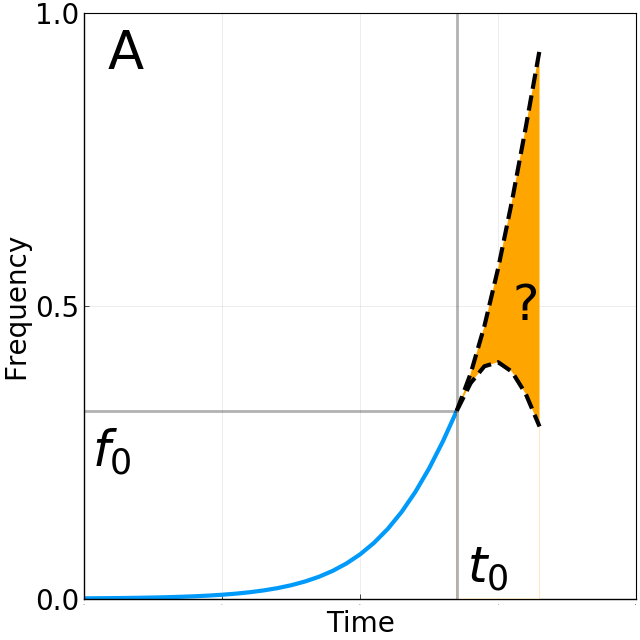
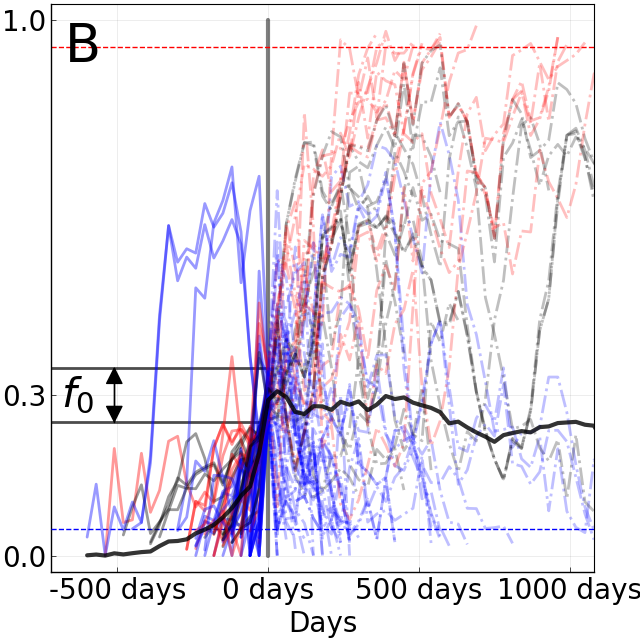
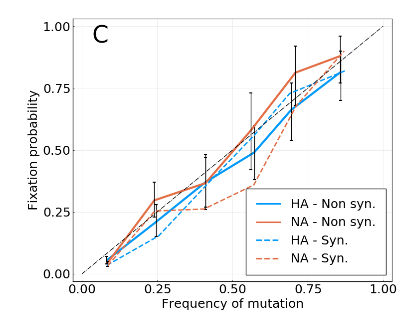
Fixation probability
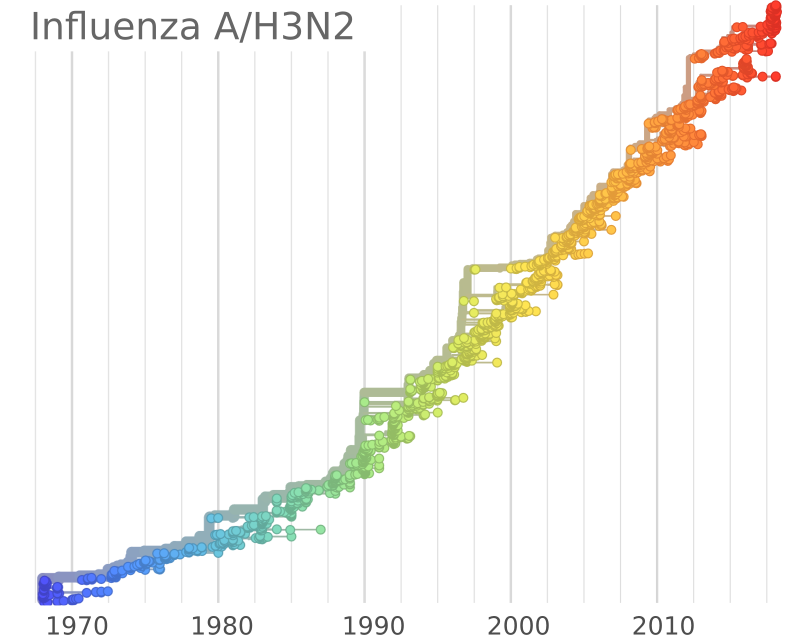
A/H3N2 influenza
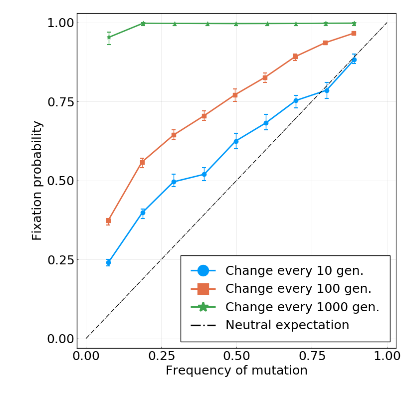
Simulations with increasing interference
Diversity in immune responses
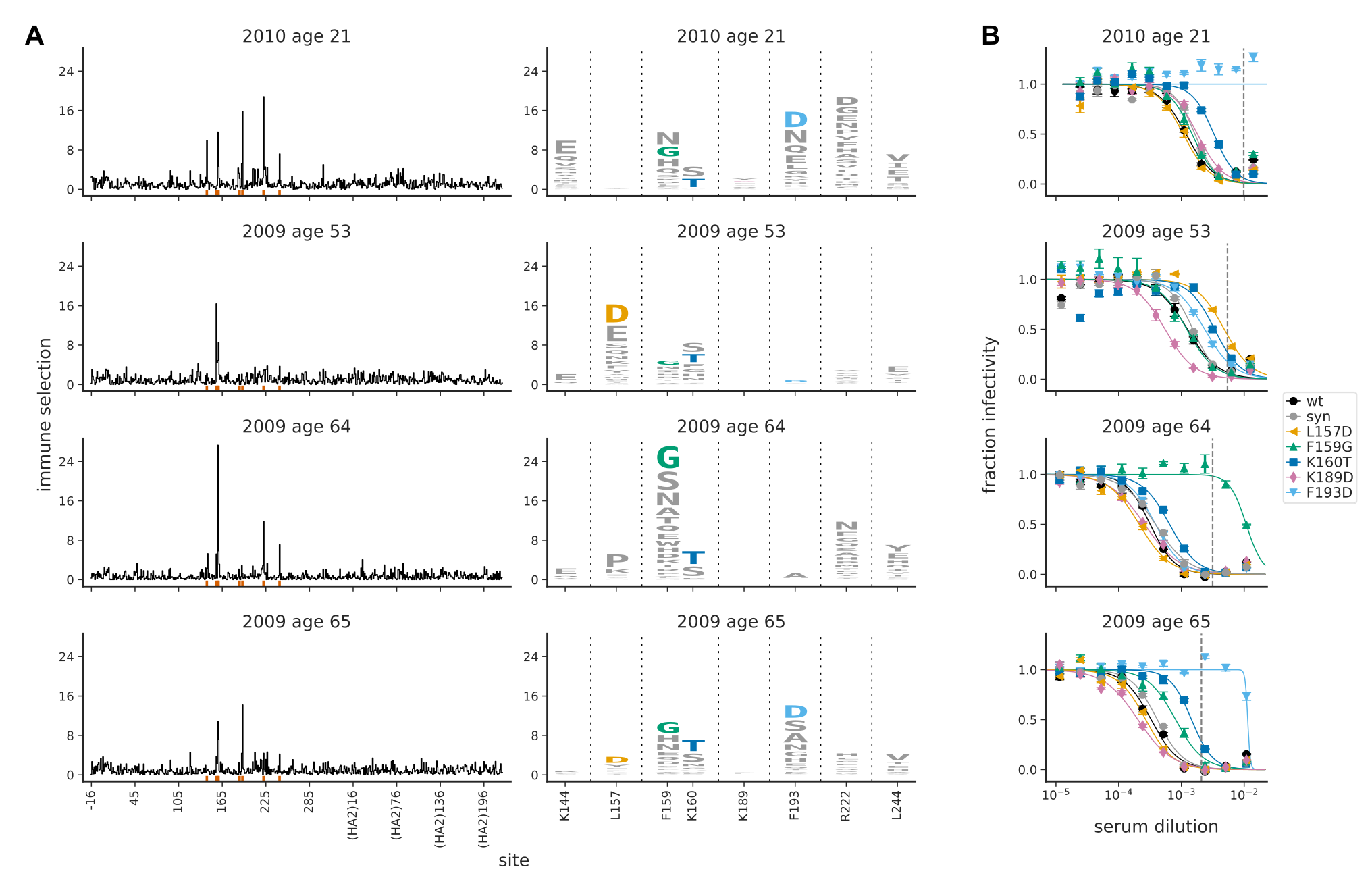
A/H3N2 influenza doesn't quite fit
- Strong signal of positive selection
- Rapid clade displacement, interference
- Any given escape only allows to escape a fraction of the population
→ expiration of selection long before a mutation is common - The more diverse the immune landscape, the more neutral it looks
- Still, evolutionary rate at antigenic sites very high
Influenza and Theory acknowledgments





- Boris Shraiman
- Colin Russell
- Trevor Bedford
- Pierre Barrat
- Oskar Hallatschek
- All the NICs and WHO CCs that provide influenza sequence data
- The WHO CCs in London and Atlanta for providing titer data



Acknowledgments
Trevor Bedford and his lab -- terrific collaboration since 2014

especially James Hadfield, Emma Hodcroft, Ivan Aksamentov, Moira Zuber, and Tom Sibley
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID


Why do predictions work