Real-time tracking of SARS-CoV-2 using Nextstrain
Richard Neher
Biozentrum & SIB, University of Basel
slides at neherlab.org/202102_abwasser.html
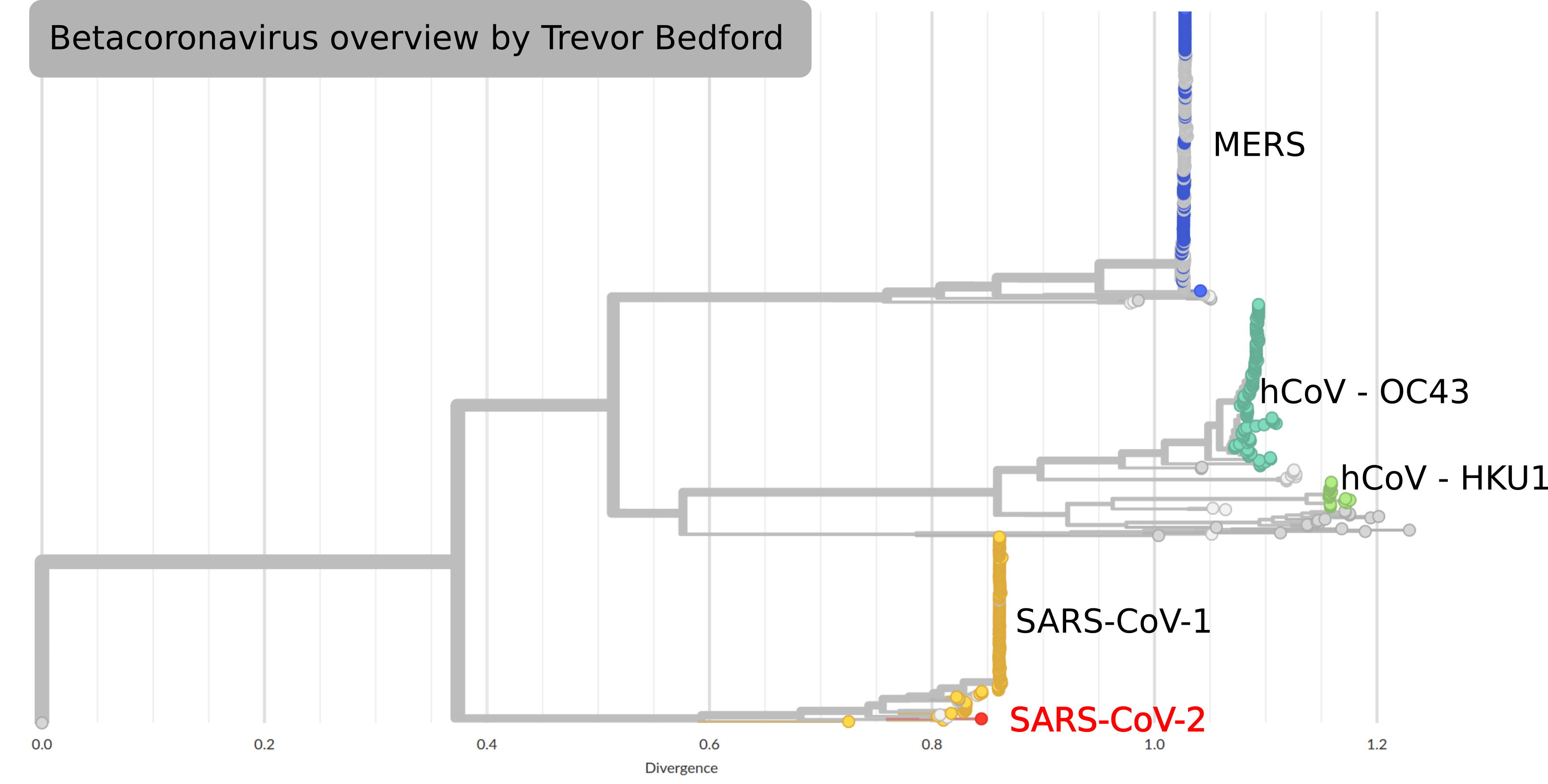 by Trevor Bedford
by Trevor Bedford
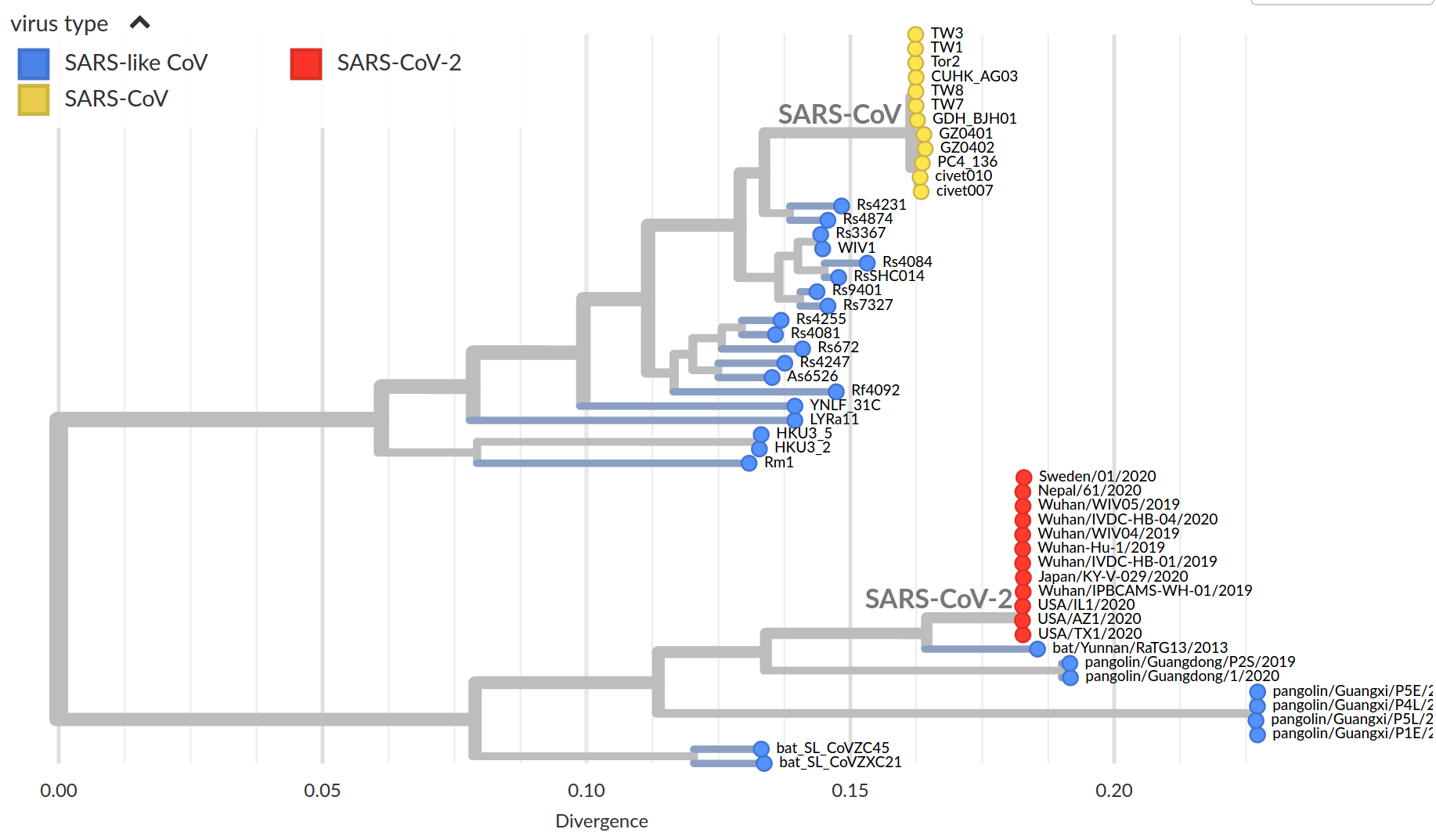 by Trevor Bedford
by Trevor Bedford
Sequences record the spread of pathogens




Tracking diversity and spread of SARS-CoV-2 in Nextstrain
Available data on Jan 26
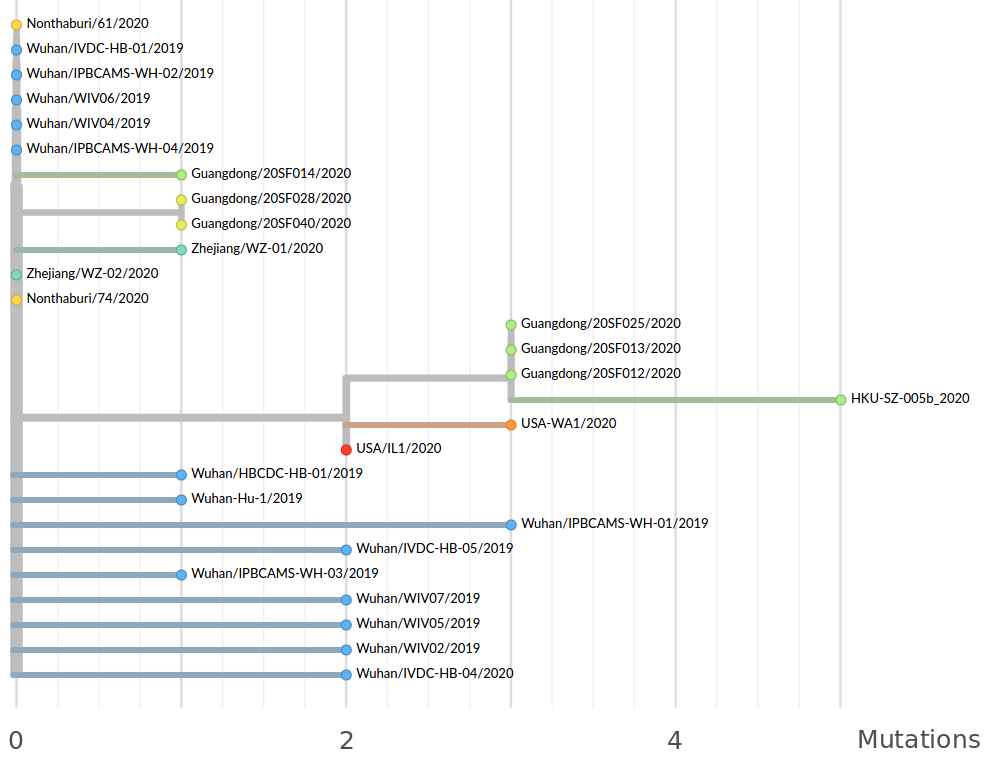
Early genomes differed by only a few mutations, suggesting very recent emergence
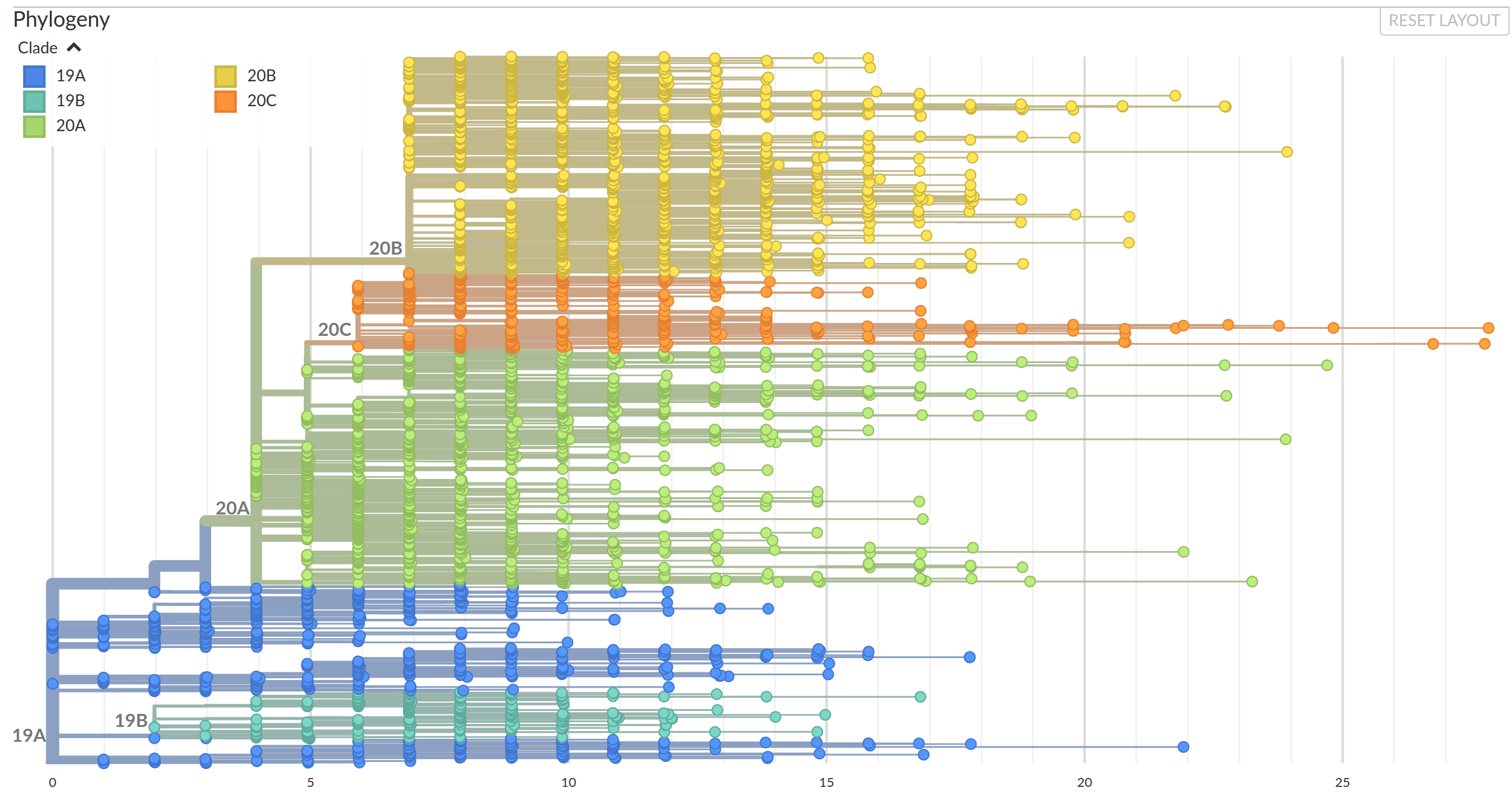
Summer 2020: Mutations accumulate constantly, but most of them are irrelevant and rare.
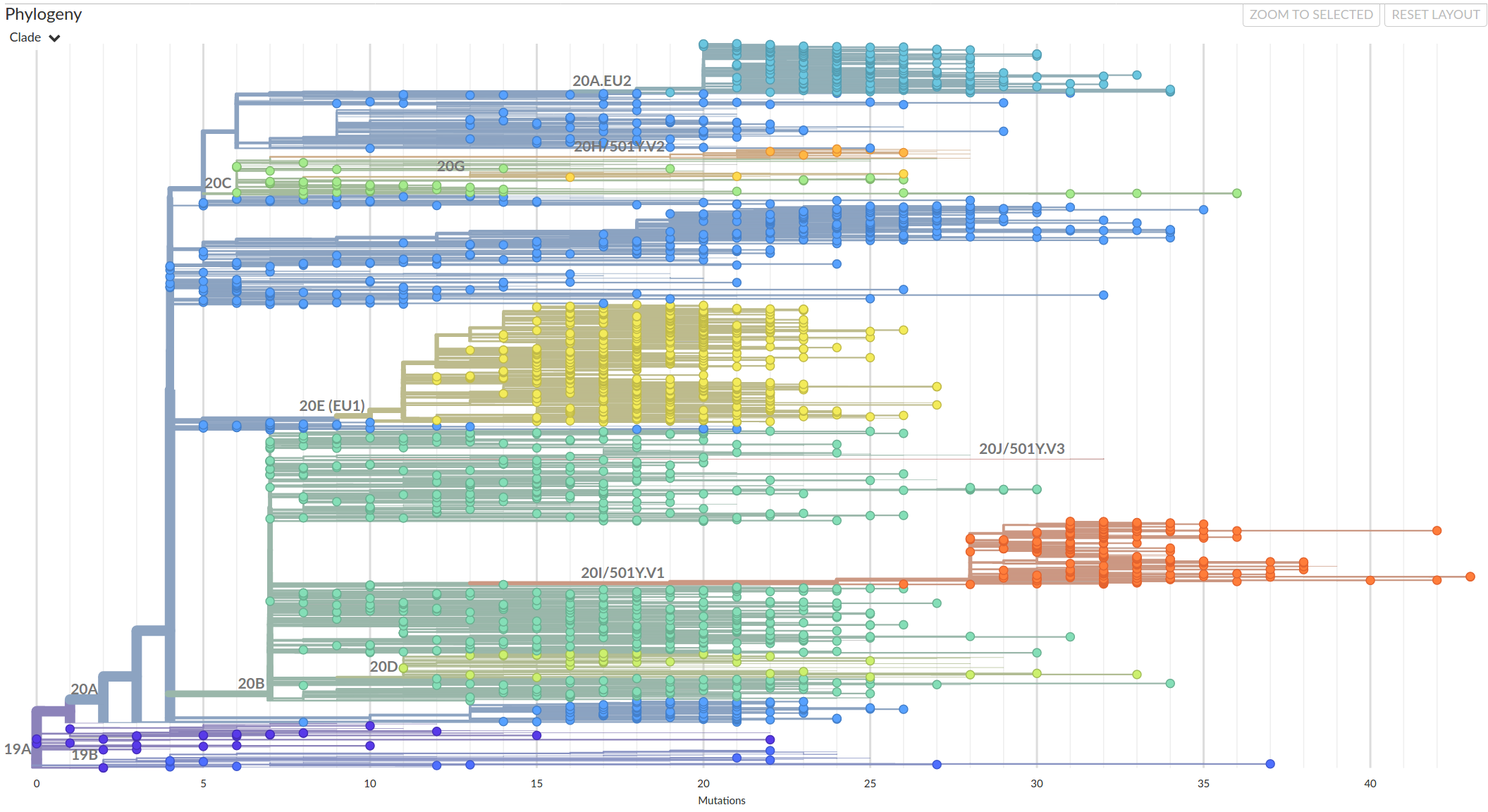
In the second half of 2020, Variants of Concern emerged.
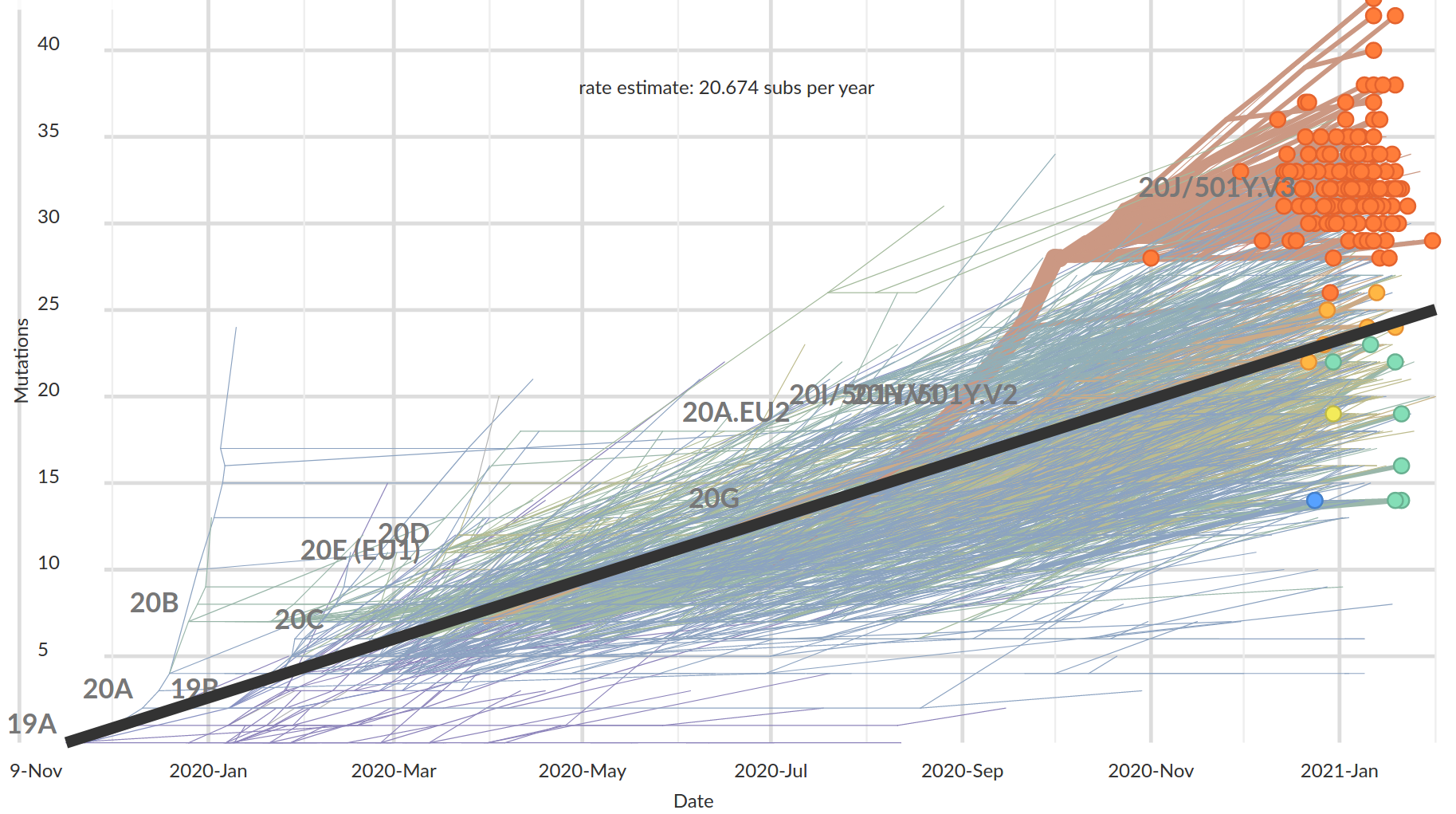
The genome accumulates about two mutations a month... but some variants are more diverged
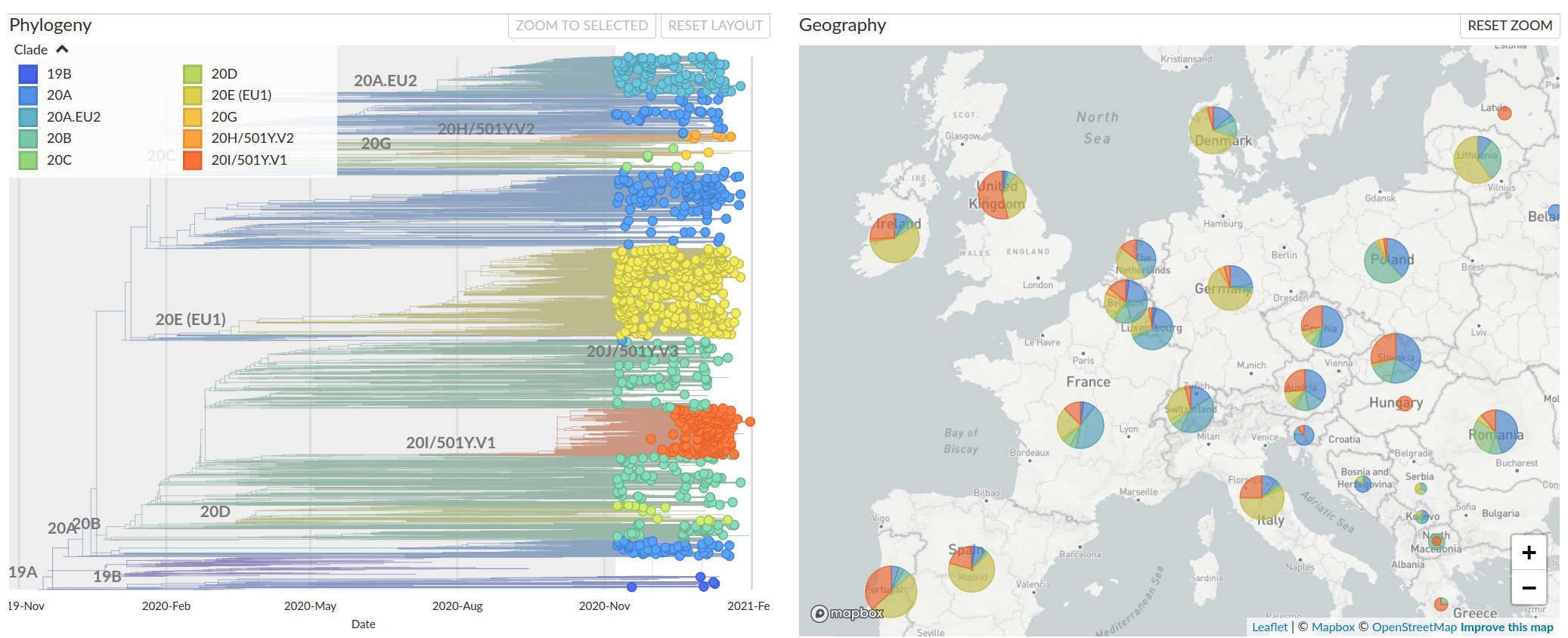
Diversification into multiple clades/variants distributed across Europe.
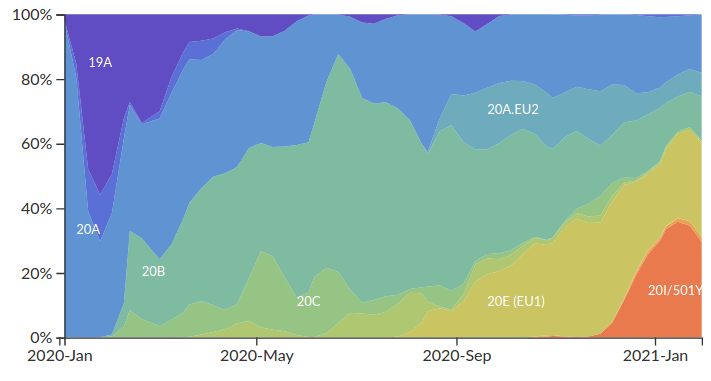
Variants are rising and falling in frequency.
Why molecular surveillance of novel variants?
- Epidemiological investigations
- Molecular correlates of changing patterns in spread and disease
- Early warning signals
- Changes in transmissibility or presentation
- Immune-escape variants and effects on vaccine efficacy
- Essential input for future vaccine strain selection process
Acknowledgments
Trevor Bedford and his lab -- terrific collaboration since 2014

especially James Hadfield, Emma Hodcroft, Ivan Aksamentov, Moira Zuber, and Tom Sibley
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID

