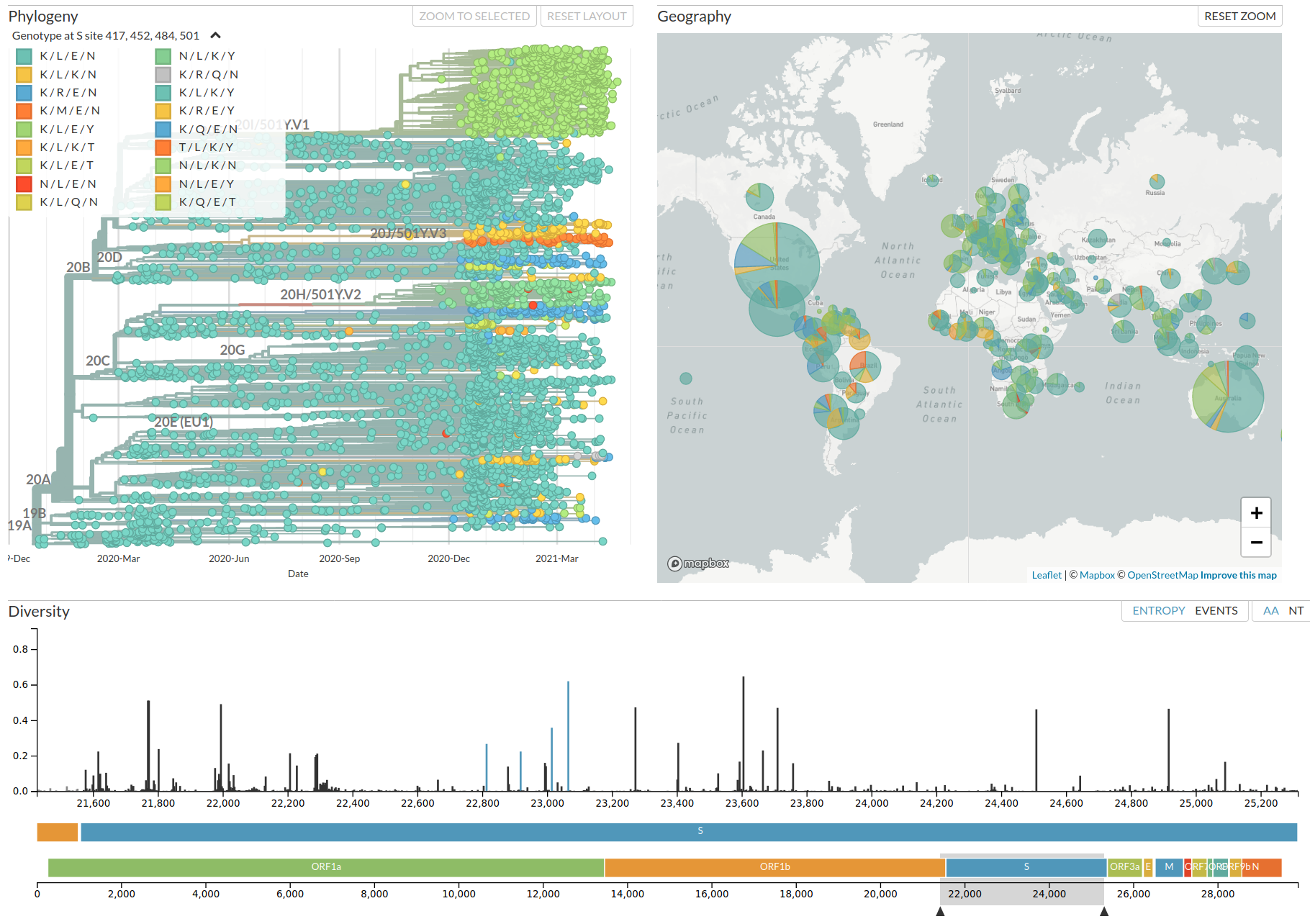
Assessing SARS-CoV-2 variants early on
Richard Neher
Biozentrum & SIB, University of Basel
slides at neherlab.org/202104_ECDC.html
Similarities to influenza virus evolution
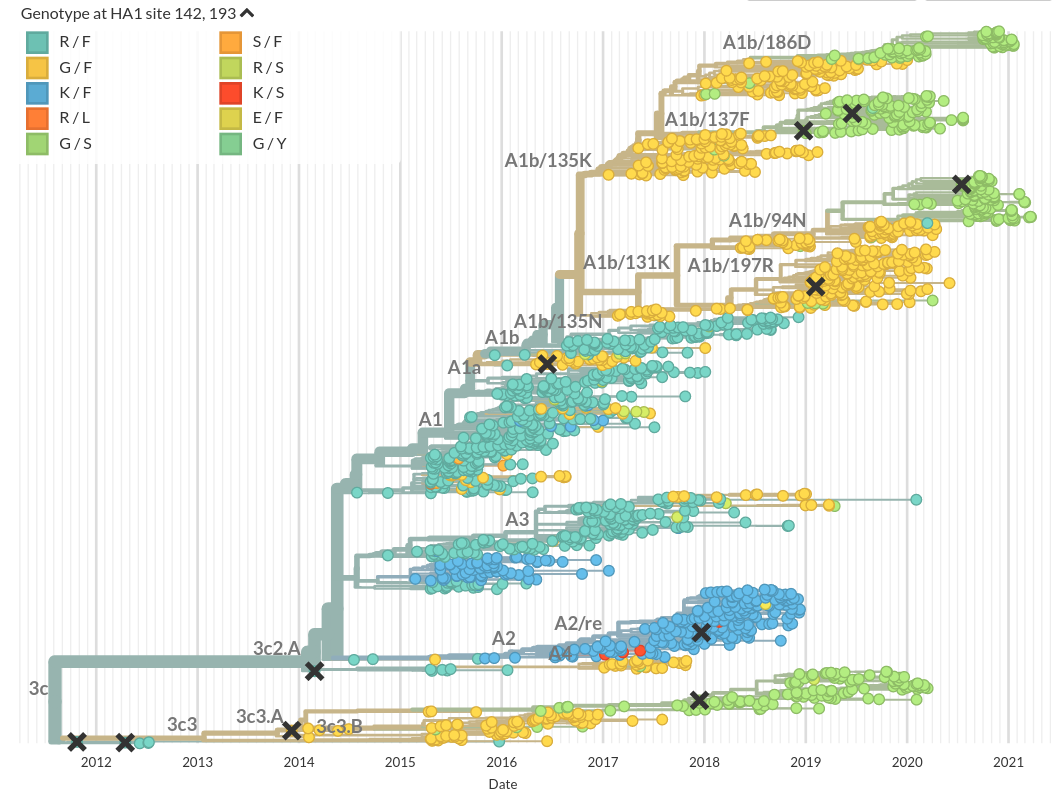
 Klingen et al, Trends in MicroBio, 2018
Klingen et al, Trends in MicroBio, 2018
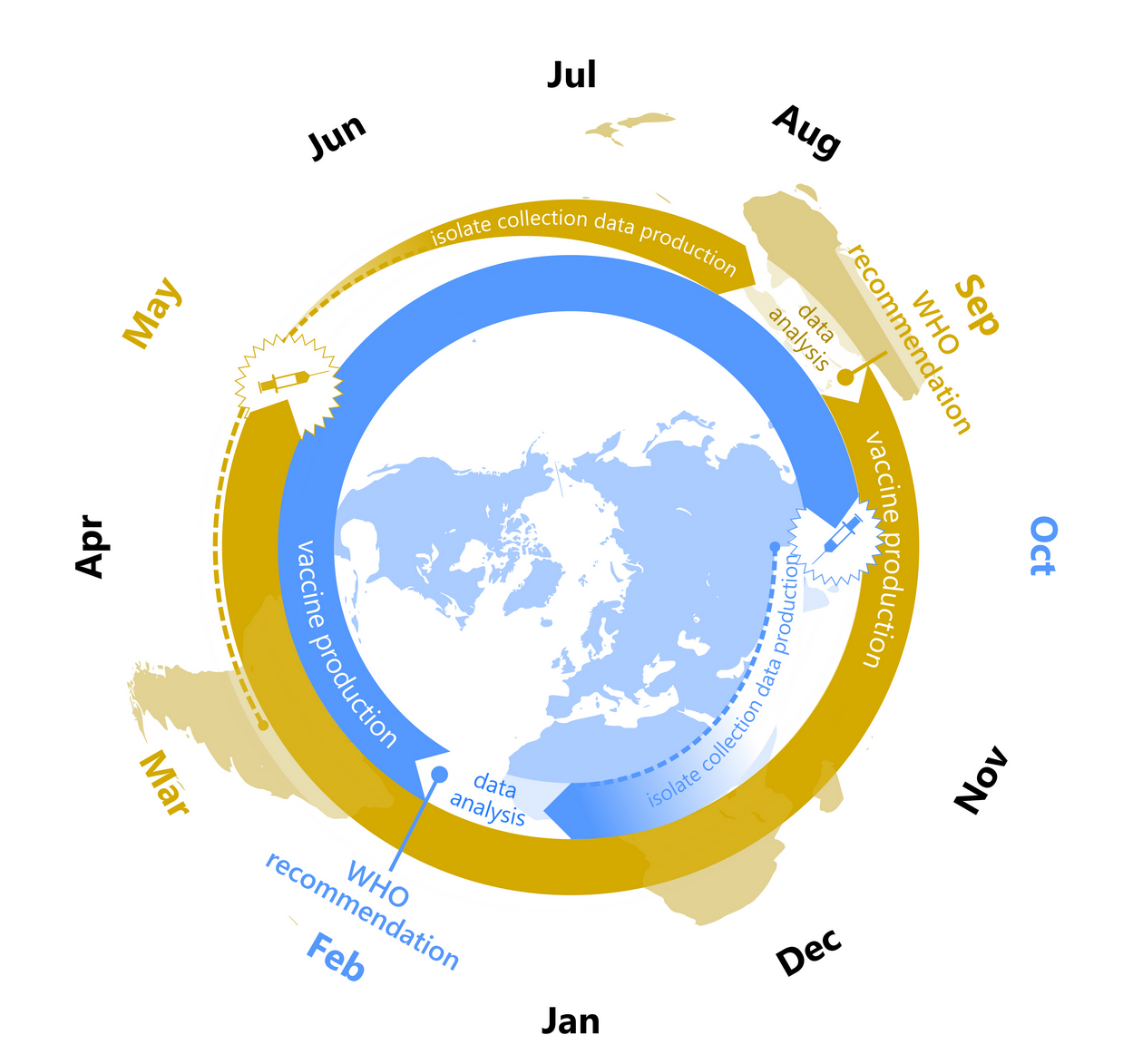
Continuous back and forth between experimental and computational groups.
Weekly sequence submissions
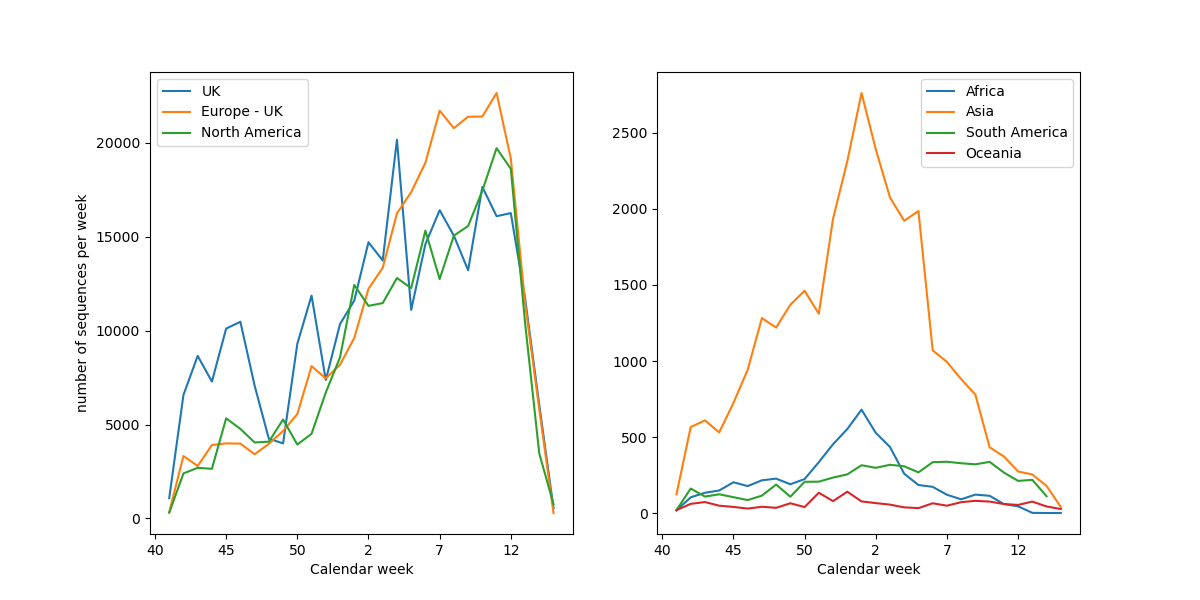
Data we analyze are contributed by scientists from all over the world
Data are shared via and curated by GISAID

Challenges assessing novel variants
- Frequency dynamics are skewed by sampling biases and varying incidence
- Rare sequences might represent large outbreaks in poorly sampled regions
- Sudden changes in sampling strategies might look like rapidly increasing frequencies
- Focus on specific mutations in known VoCs might miss important others
- Laboratory characterization takes time
- Prioritization of variants for laboratory characterization.
- Harmonized categorization of sequences as
- General surveillance
- Outbreak investigations
- Vaccine breakthroughs, reinfections, etc
- Travel associated cases
- Support for denser sampling across the globe
(some places in Europe are still undersampled)
We need a sustainable and coordinated approach.
- Avoid too much duplication of efforts
- Maximize usefulness and usability of data and analyses
- Establish exchange between groups with different expertise and stakeholders
Deep mutational scanning for RBD mutations that affect AB binding and ACE2 binding
- By Tyler Starr, Allie Greaney, Jesse Bloom and colleagues
- Systematic evaluation of all RBD mutations for escape and ACE2 binding
- Monoclonals binding to class 1,2,3,4 epitopes
- Convalescent and vaccine sera to map population escape
- Escape maps of indivual sera and antibodies by Bloom lab
- Escape calculator for retained binding by Jesse Bloom
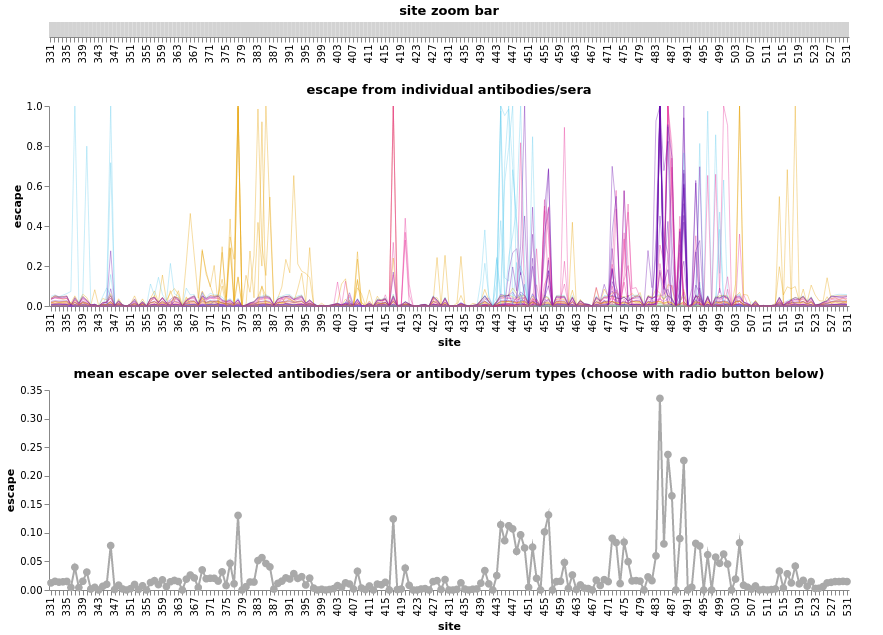
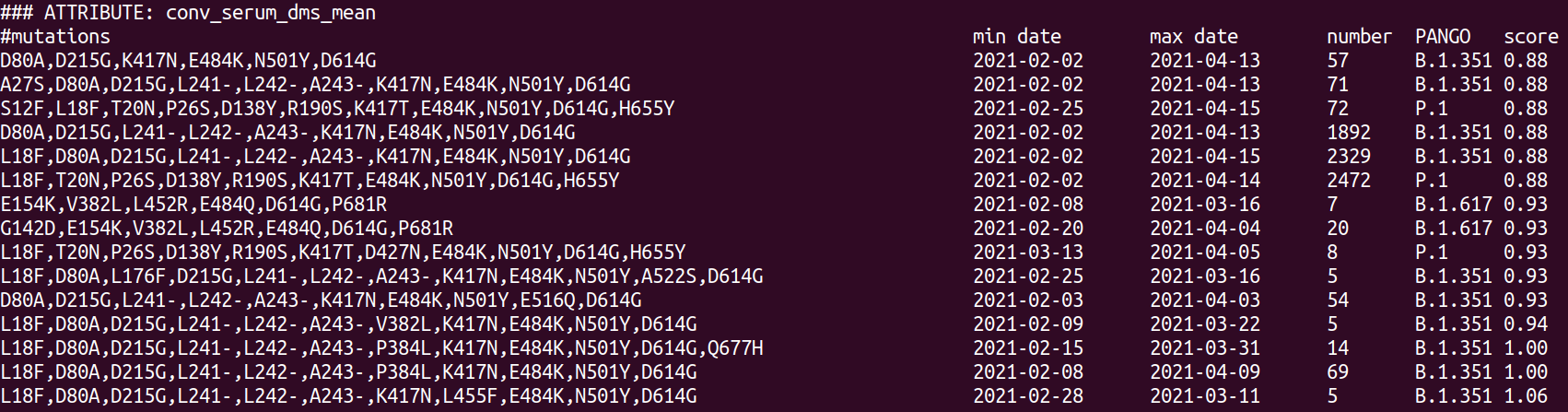
Scoring all sequences for further characterization
- Escape in distinct RBD epitope classes
- RBD escape in convalescent/vaccine sera
- NTD mutations
- Other VoC associated mutations (nsp6 del)
- ACE2 binding
- Furin cleavage site mutations
- Recent growth
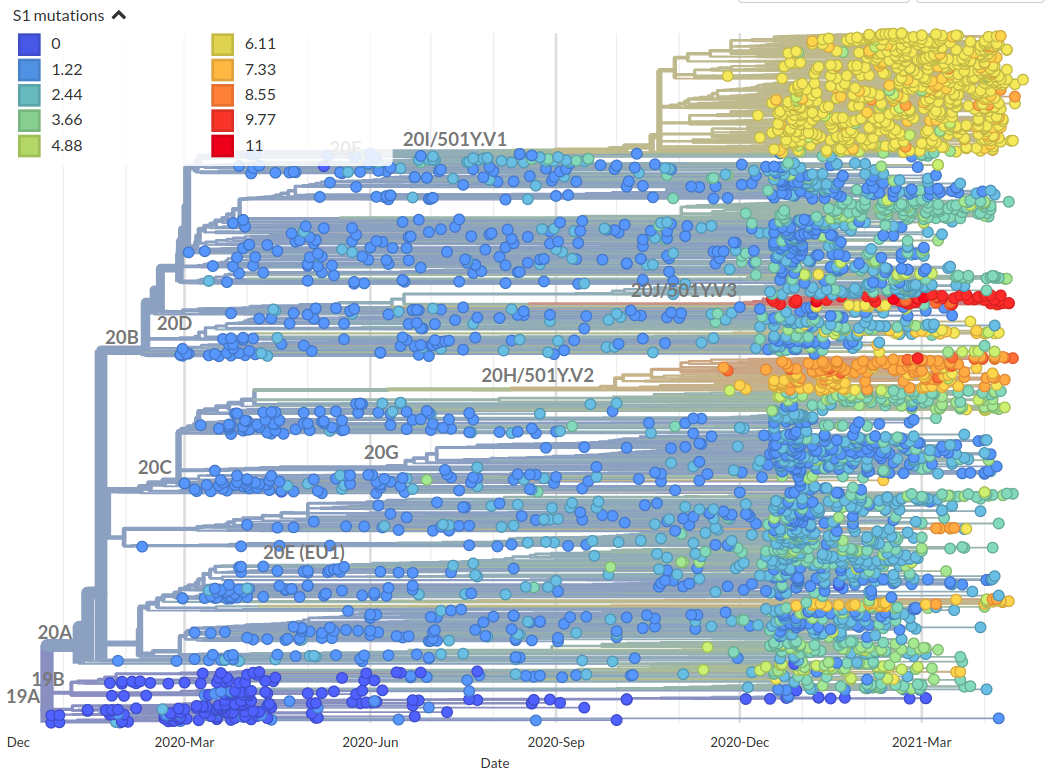
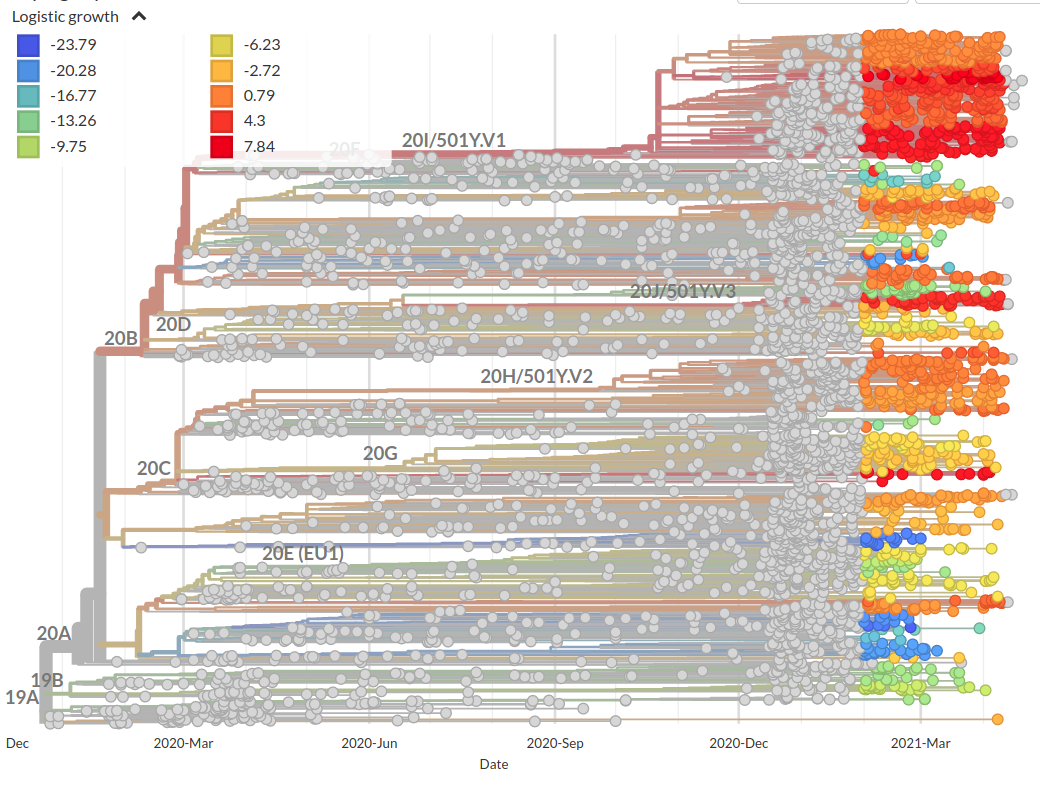
Incorporation of scores into phylogenetic analysis
Acknowledgments
Trevor Bedford and his lab -- terrific collaboration since 2014

especially James Hadfield, Emma Hodcroft, Ivan Aksamentov, Moira Zuber, and Tom Sibley
Allie Greaney and Jesse Bloom for discussion of the dms data
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID

