Tracking and predicting the evolution of human RNA viruses
Richard Neher
Biozentrum & SIB, University of Basel
slides at neherlab.org/202104_Karolinska.html
Human seasonal influenza viruses

Positive tests for influenza in the USA by week
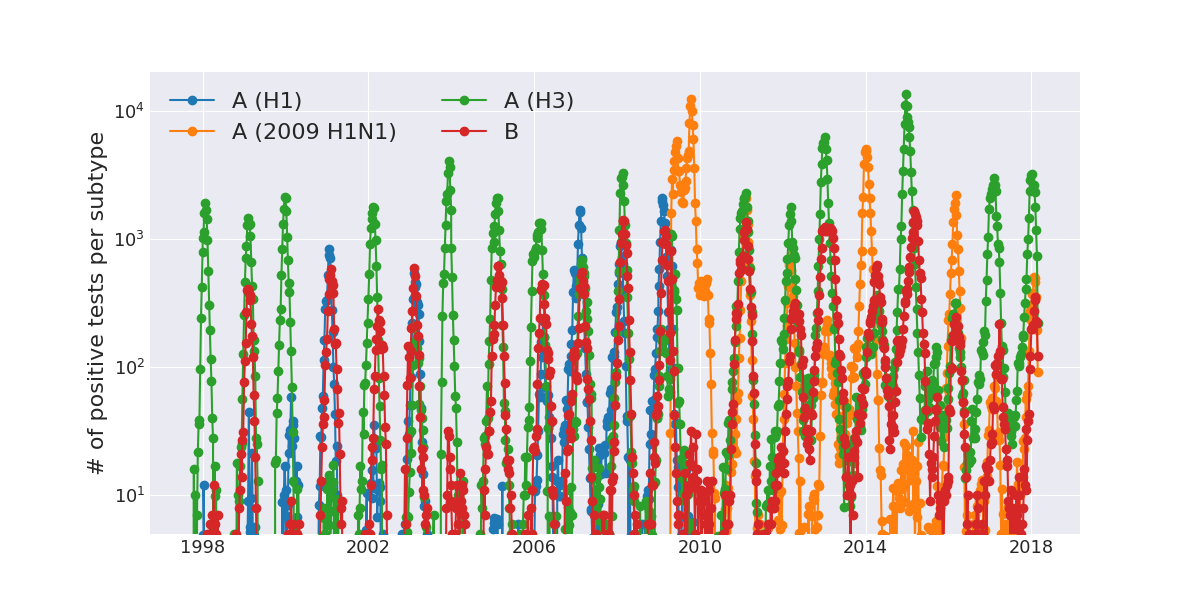 Data by the US CDC
Data by the US CDC
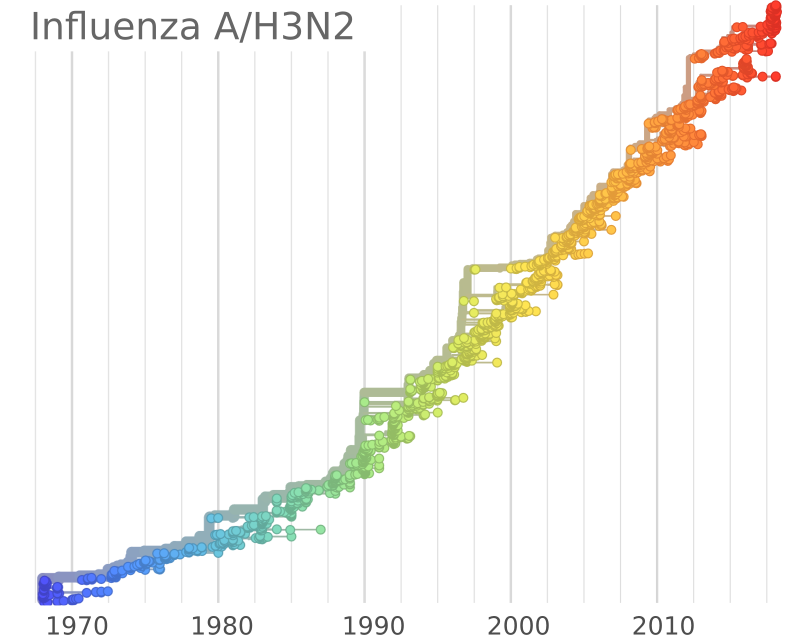
Sequences record the spread of pathogens




Mutations accumulate at a rate of $10^{-5}$ per site and day!



- Influenza viruses evolve to avoid human immunity
- Vaccines need frequent updates

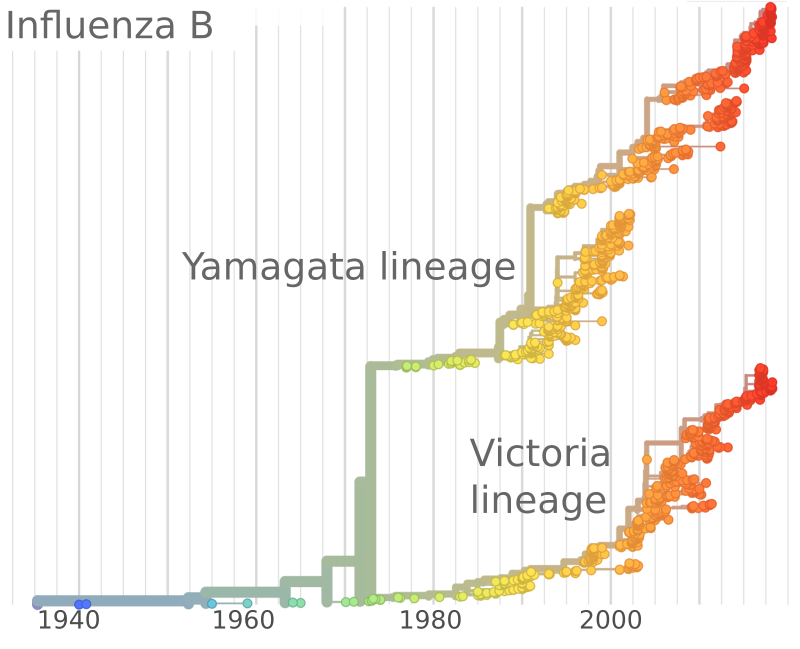
Influenza B viruses have split into two lineages
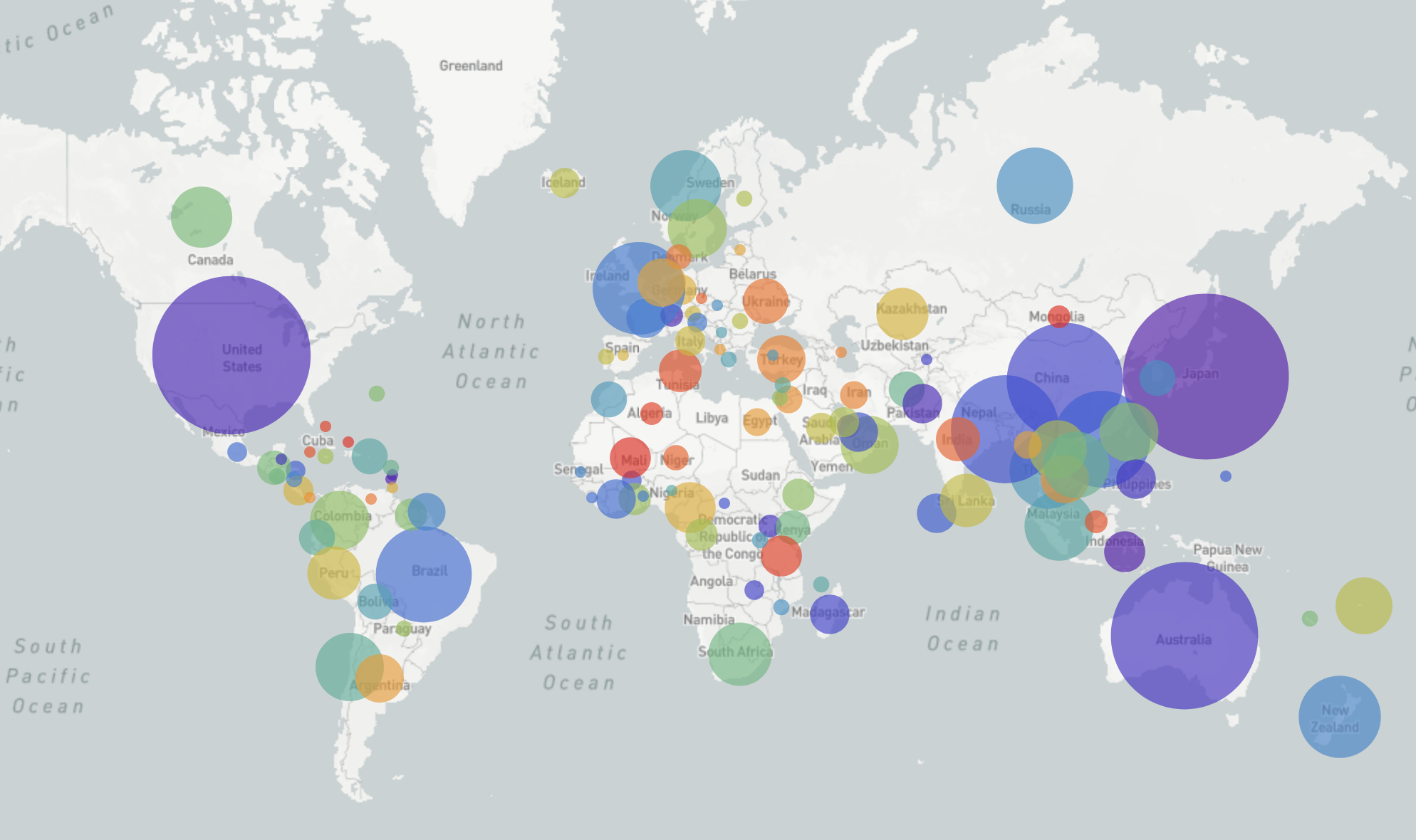
GISRS and GISAID -- Influenza virus surveillance
- comprehensive coverage of the world
- timely sharing of data through GISAID -- often within 2-3 weeks of sampling
- hundreds of sequences per week (in peak months)
→ requires continuous analysis and easy dissemination
→ interpretable and intuitive visualization
nextflu.org
joint project with Trevor Bedford & his lab
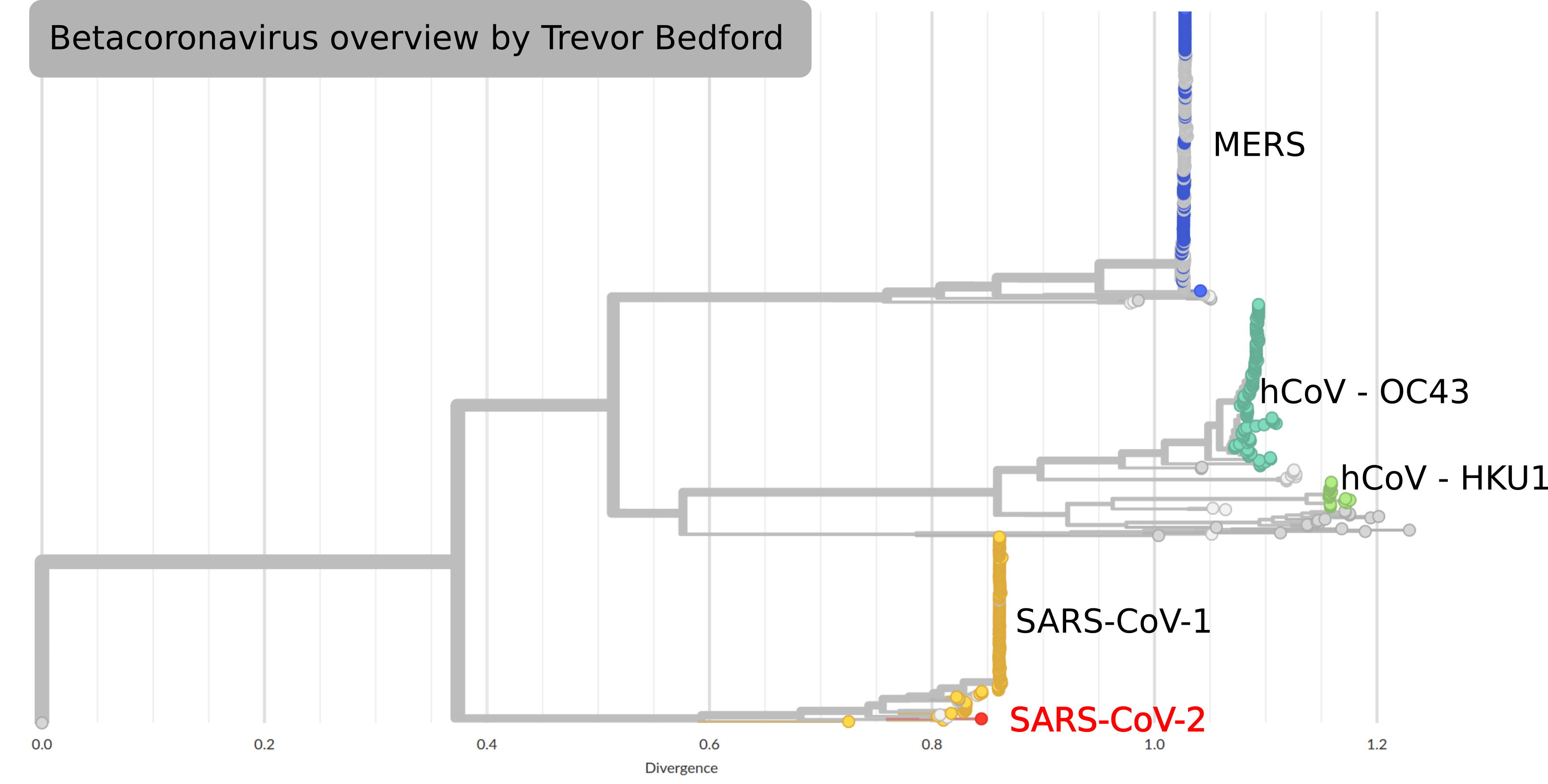 by Trevor Bedford
by Trevor Bedford
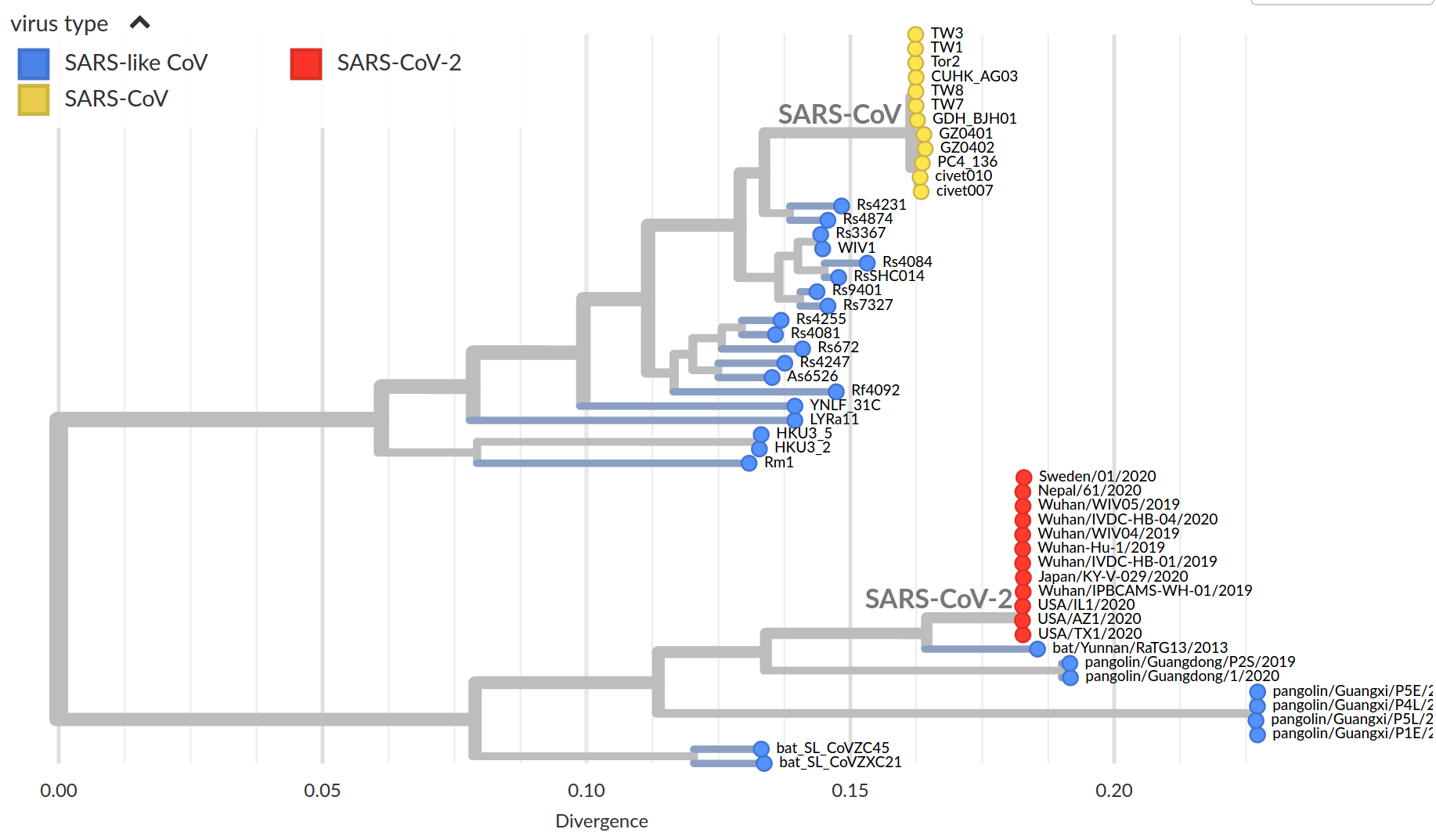 by Trevor Bedford
by Trevor Bedford
Tracking diversity and spread of SARS-CoV-2 in Nextstrain
Available data on Jan 26
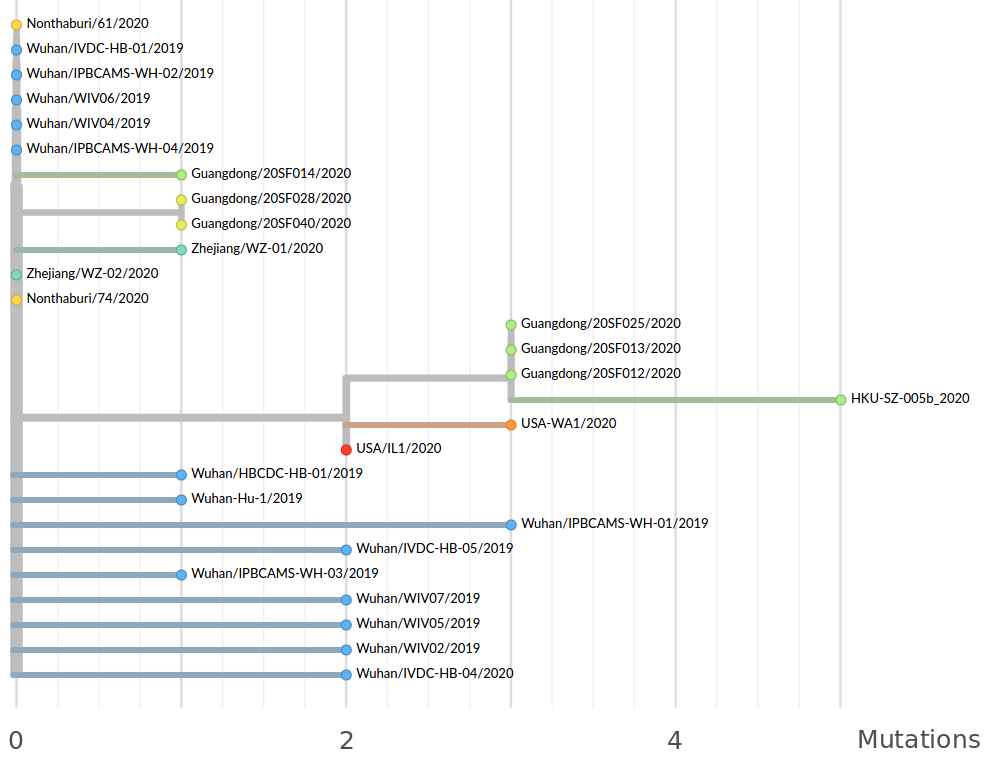
Early genomes differed by only a few mutations, suggesting very recent emergence
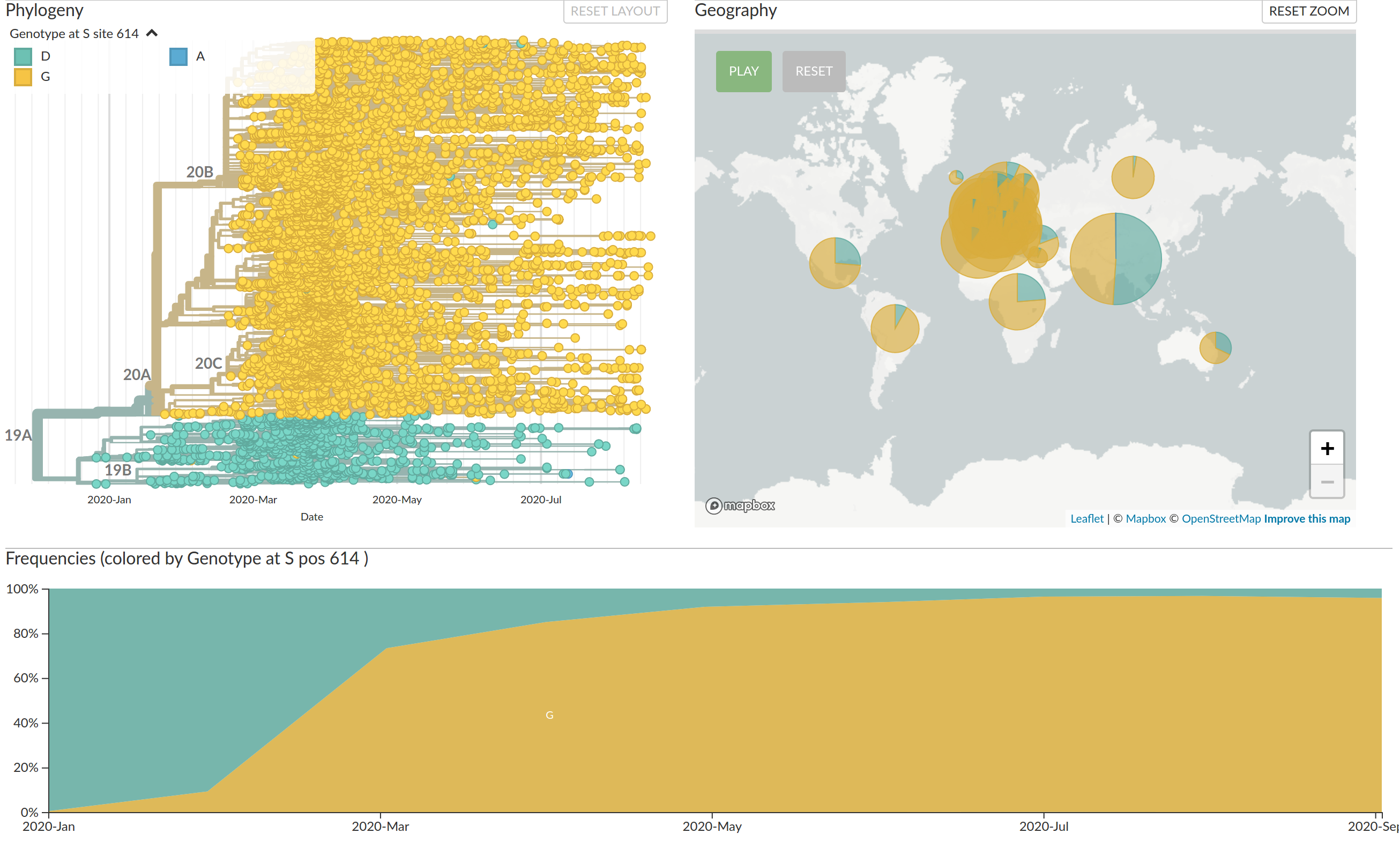
Interactive part on Nextstrain
VoCs have more mutations than expected...
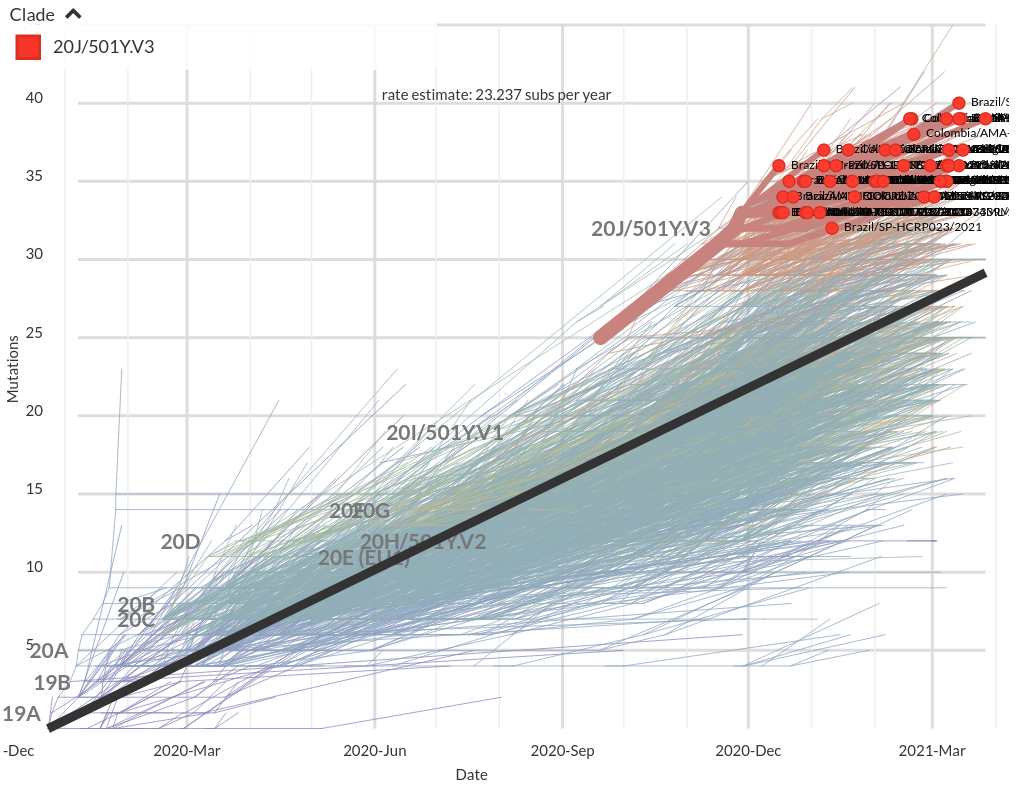 nextstrain
nextstrain
VoCs and VoIs: rapid converging evolution
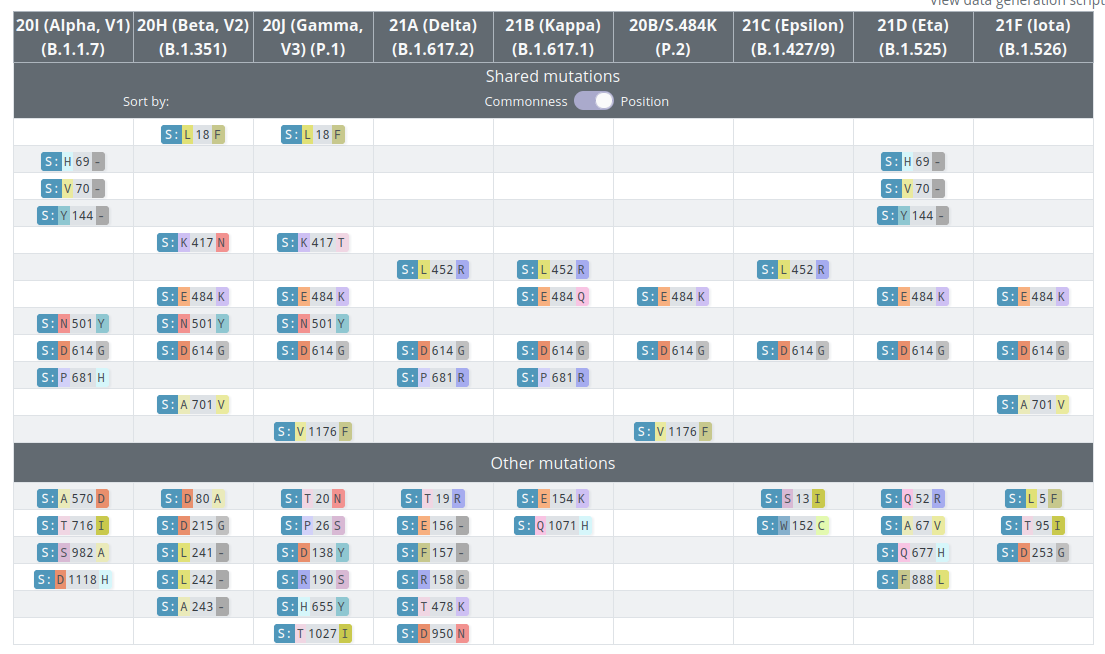 covariants.org by Emma Hodcroft
covariants.org by Emma Hodcroft
Reduced neutralization of VoCs by convalescent serum (B.1.351)
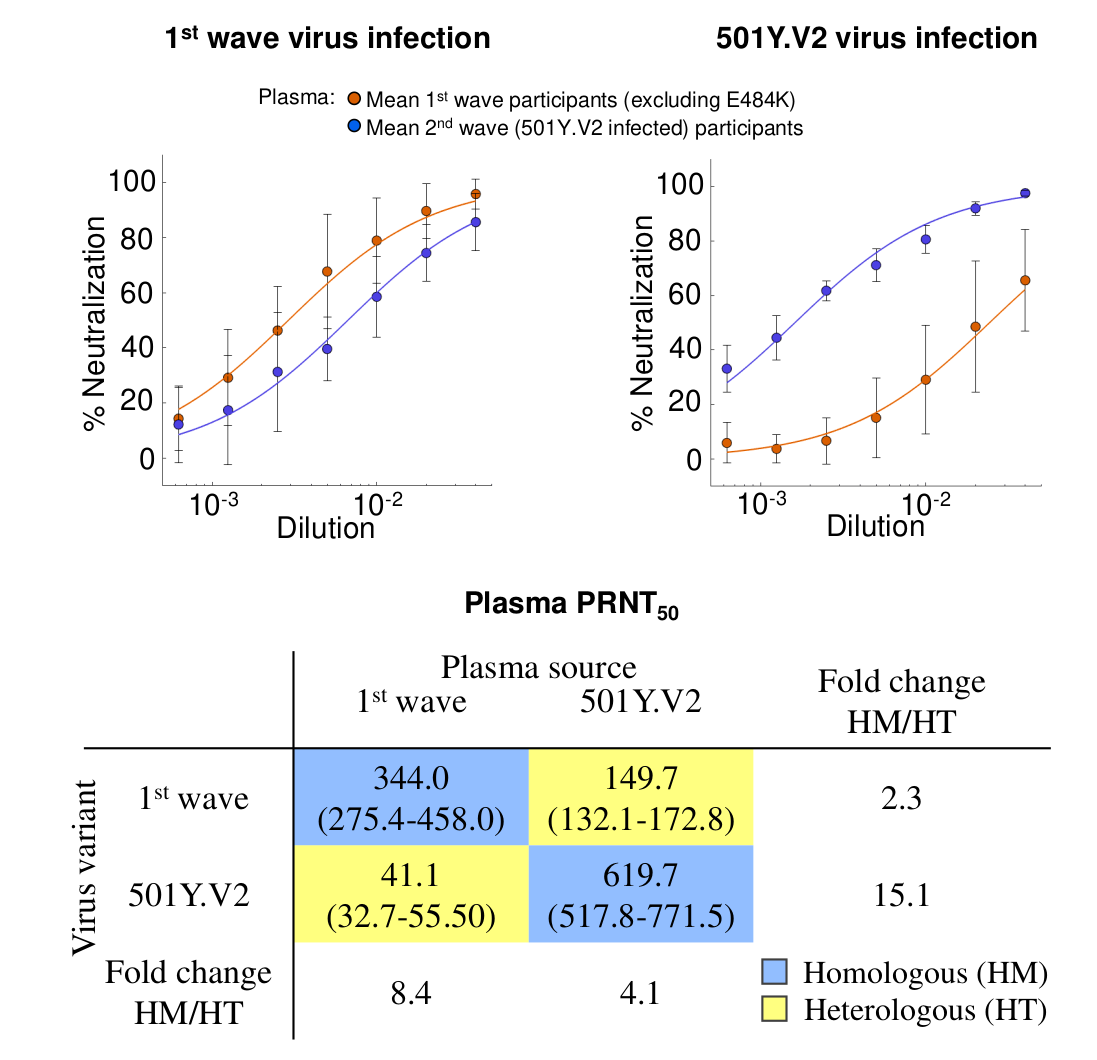
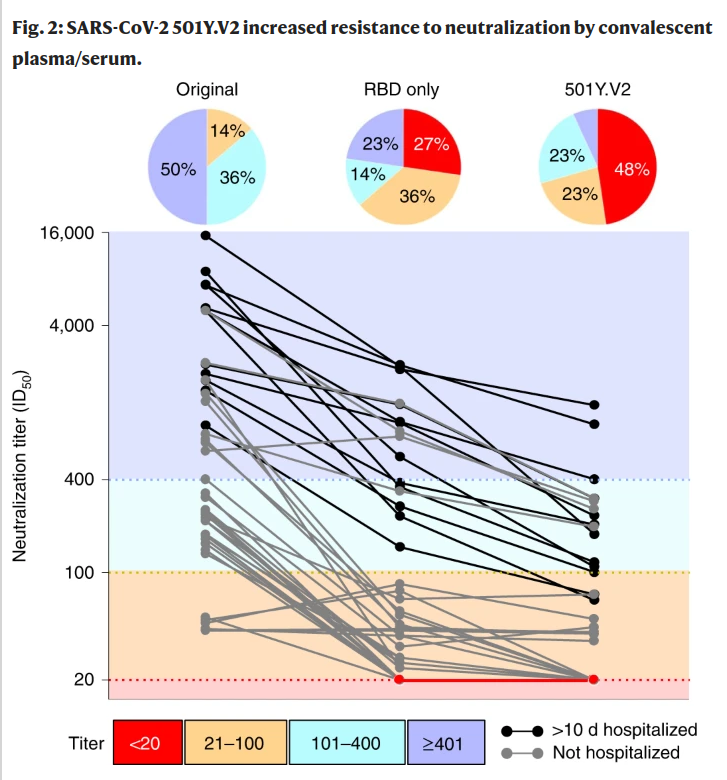
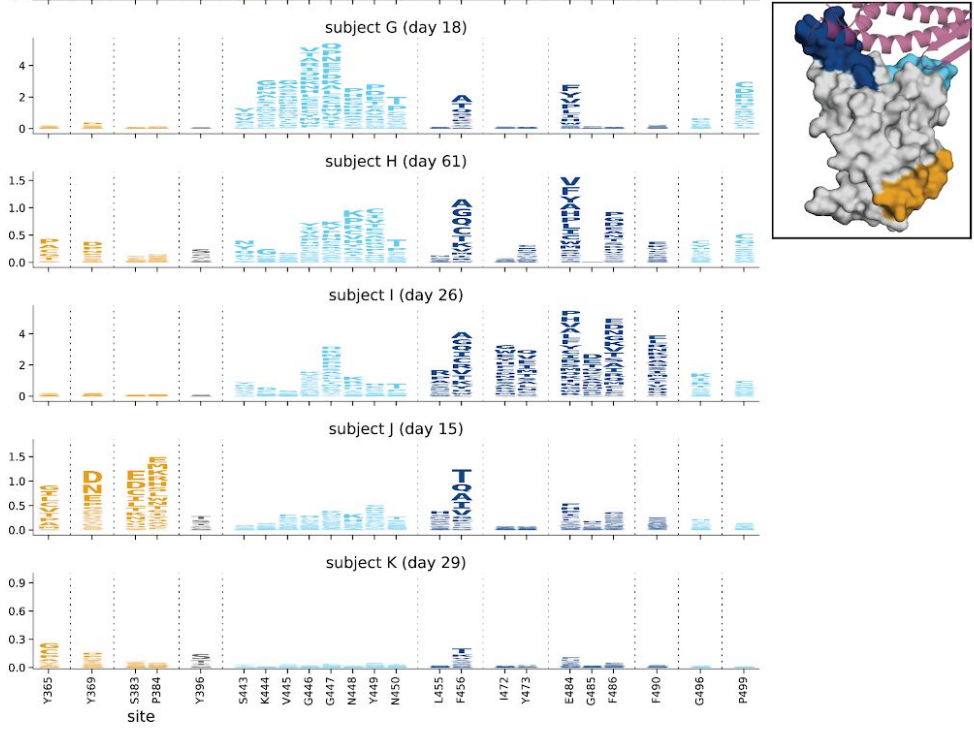
Deep mutational scanning approaches
- Library of pseudo-typed virus with mutations at every position
- Propagate in cell culture with and without selection (serum, ABs)
- Measure which variants are selected for
- → assess mutations before they are observed
- → screen for potentially important variants
- → follow-up with virological/serological studies
Summary and Conclusions
- Enormous amounts of data provide unprecedented real time picture of a pandemic
- Several highly evolved variants of SARS-CoV-2 emerged in a short time
- The current VoCs are likely just the tip of the iceberg
- Processing, analyzing, and interpreting data is a major challenge
- Integration of genomic and experimental data is key to generate insight
Acknowledgments
Trevor Bedford and his lab -- terrific collaboration since 2014

especially James Hadfield, Emma Hodcroft, Ivan Aksamentov, Moira Zuber, and Tom Sibley
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID


Influenza and Theory acknowledgments





- Boris Shraiman
- Colin Russell
- Trevor Bedford
- Pierre Barrat
- Oskar Hallatschek
- All the NICs and WHO CCs that provide influenza sequence data
- The WHO CCs in London and Atlanta for providing titer data


