SIB resource candidate: Nextstrain
Richard Neher
Biozentrum & SIB, University of Basel
slides at neherlab.org/202104_SIB_resource.html
Origins of Nextstrain
- Started as an influenza specific project in 2015
- Meant to accompany attempts to predict future influenza population with "always up-to-date" trees and predictions
- Initially a project of Trevor Bedford and myself
- Expansion beyond influenza (Ebola, Mers, Zika)

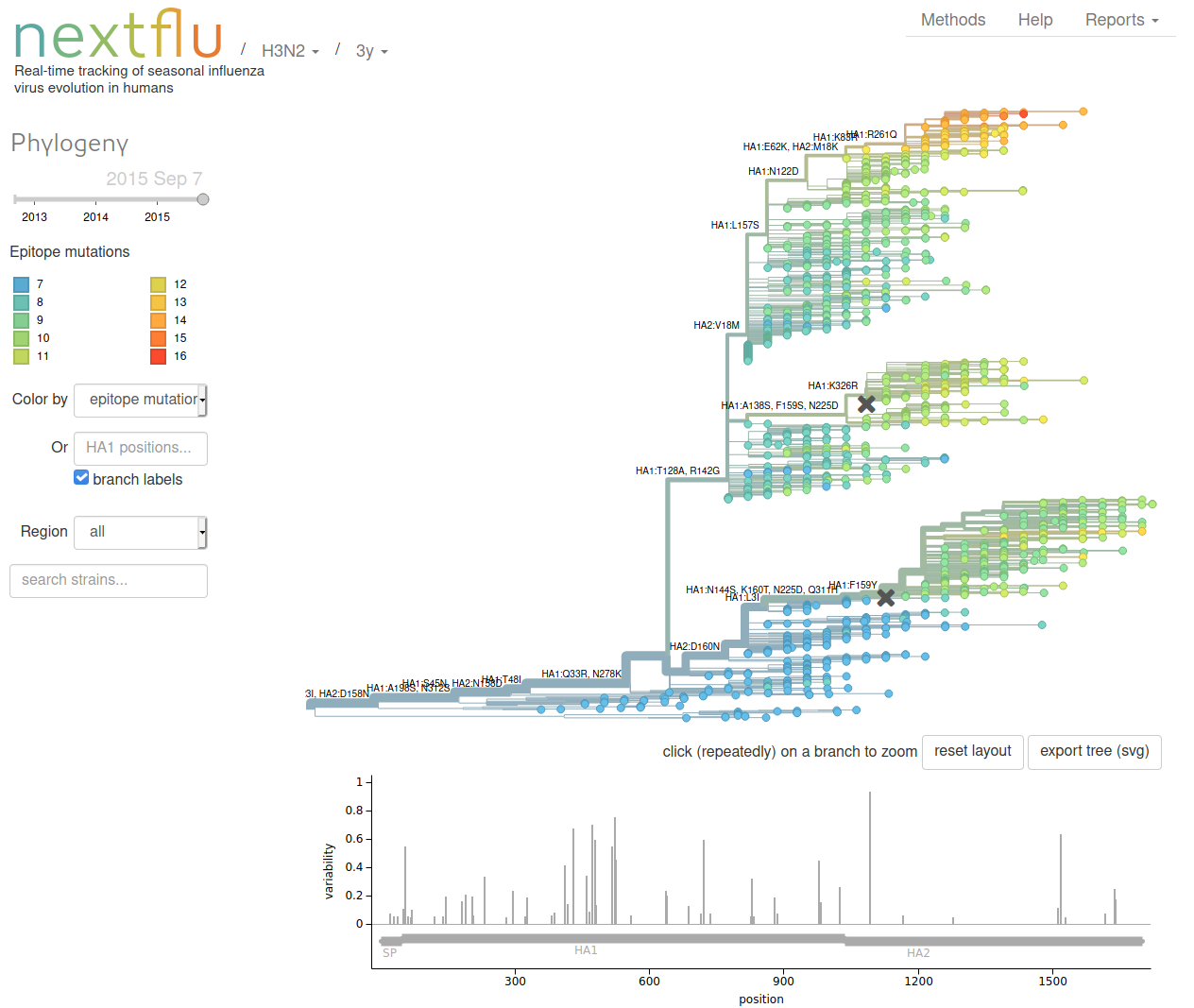
Nextstrain is both a tool chain and a web-app
Core tools by nextstrain
-
Augur -- extensible phylodynamics pipeline
- composible, modular, and extensible
- wrappers for alignment, tree building, etc
- phylodynamic and phylogeographic modules
- custom modules for frequency and fitness estimation
- Auspice -- interactive visualization
- Tree viewer, map module, flexible frequencies, charts, filters...
- Customizable and available for white labeling
- powers nextstrain.org
- of course completely open
- workflows and data (to the extend that we are allowed) open
- extensive documentation
Analyses maintained by the Nextstrain
Nextstrain-hosted community resources
Community builds via github

SARS-CoV-2 analyses by the community
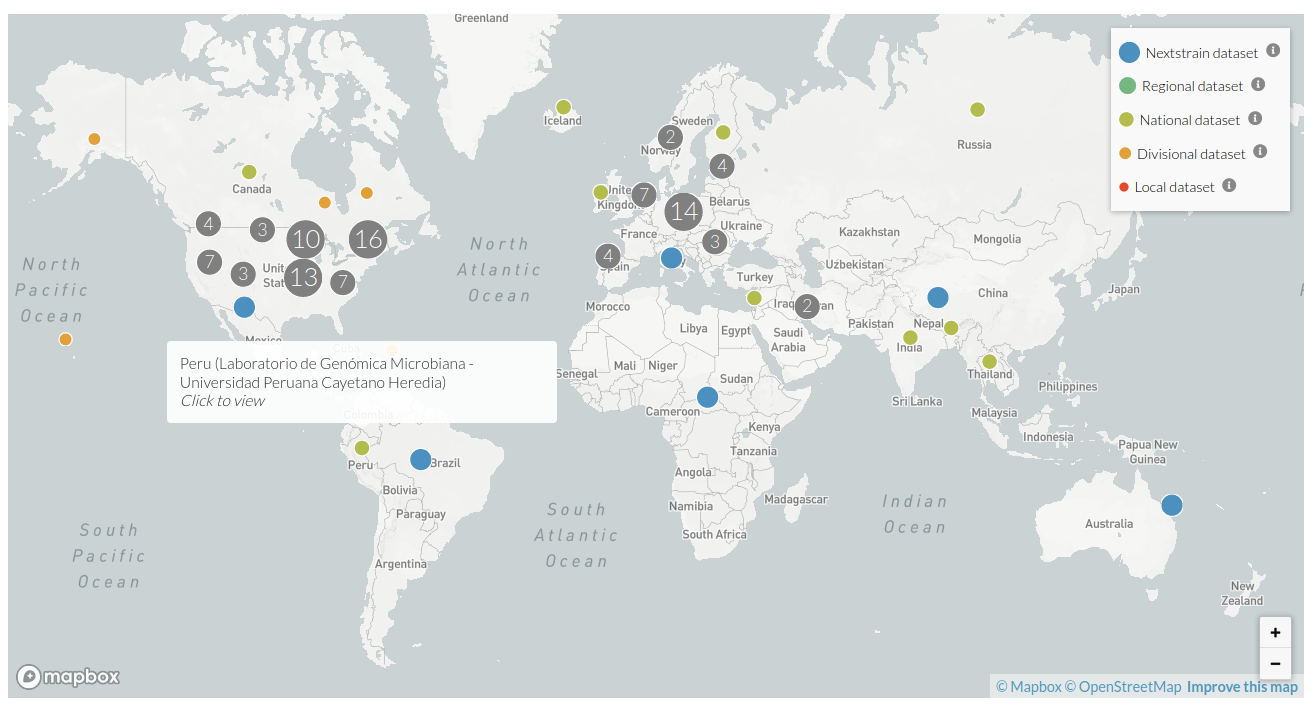
Nextstrain in the pandemic
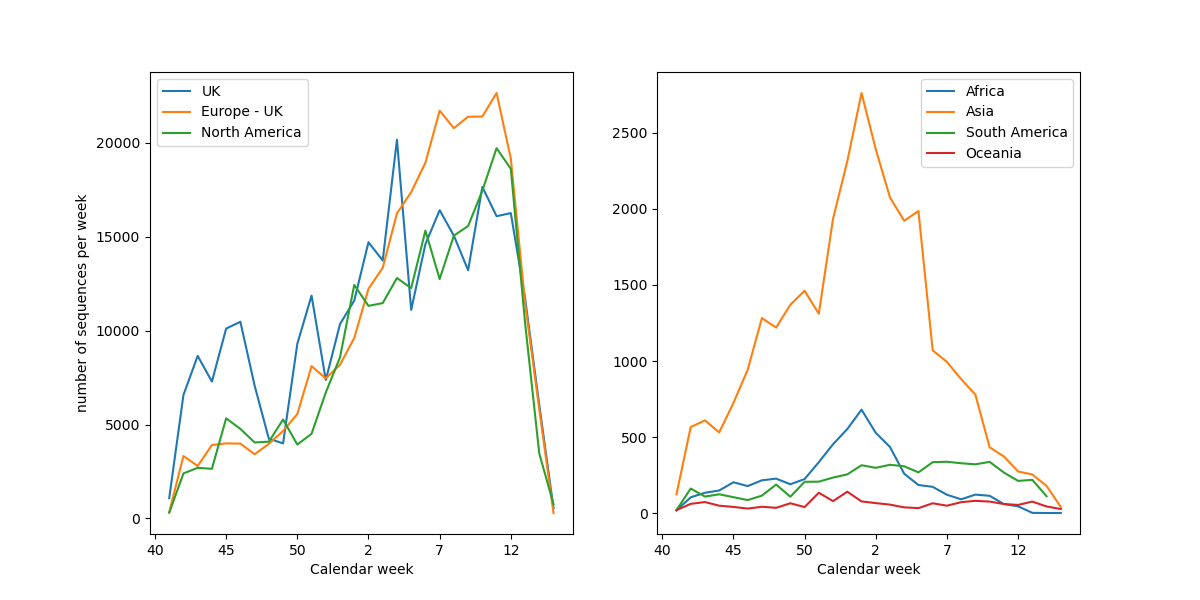
- ever increasing data volumes
- massive public interest and media coverage
- rapidly shifting requirements
Usage in Public Health and Academia

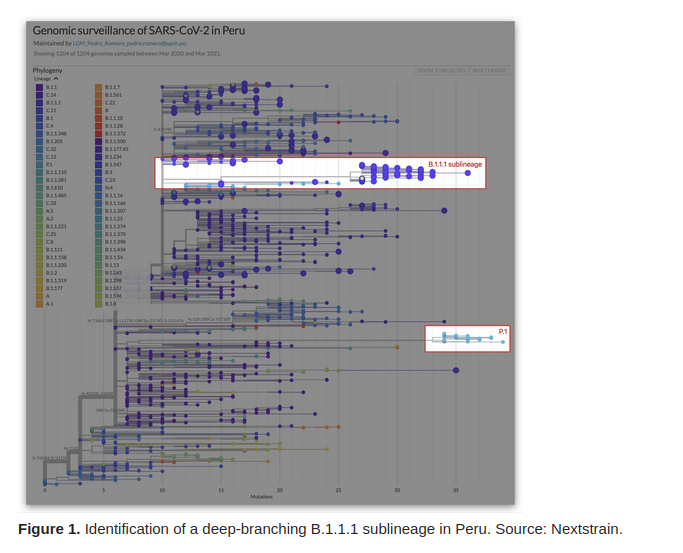
In-house use by public health stake holders
- US CDC
- Robert-Koch-Institut
- RIVM -- Dutch public health institute
- ECDC
- GISAID
- Seq-COVID Spain
- Broad Institute
- Chan Zuckerberg Institute
- and many more...
Enabling other tools
- CoVariants.org by Emma Hodcroft
- SPSP by SIB
New developments
Plans and outlook
Basel focussed efforts
-
Nextclade
- Effortless first analysis of new data
- Client based and 100% privacy preserving
- Speed-up via RUST and WASM
- Extend to other viruses
- TreeTime refactor
- workhorse of much of augur
- computational bottleneck
- huge scope for algorithmic and implementation improvements
- Reassortment, recombination, and other non-tree-like evolution
Joint or Seattle focussed efforts
-
Cloud computing
- Allow users to run their analysis via a web interface
- Host user spaces
- Integrated with Pathogen Genomics initiative of the Gates Foundation
- auspice and nextstrain.org
- More visualization components
- Data set discovery options
- Documentation
- Pathogen specific dashboards
- Integration with serological and high throughput data
Basel
-
Ivan Aksamentov (Software Engineer)

- Nextclade
- Nextclade
- CoVariants
- Covid19-Scenarios
- Bioinformatician (N.N.)
- Emma Hodcroft (former post-doc, now Bern)
- Moira Zuber (Student assistent, running analyses)
- Benjamin Otter (Student assistent, augur dev)
- Research led by myself involving 2FTEs
Seattle/NZ
- Trevor Bedford (PI)
- Tom Sibley (Software engineer)
- John Huddleston (former PhD Student/research scientist)
- James Hadfield (Software/frontend)
- Jover Lee (bioinformatics)
- Eliah Harkins (frontend/builds)
Basel
- Uni Basel/Biozentrum core funding
- Open Science Prize 2017
- Special SARS-CoV-2 call from the SNF (joint grant with Bern)
- Future: funding from BAG through WGS Surveillance initiative
Seattle/NZ
- Open Science Prize 2017
- Research grants
- Philantropy
