Virus evolution and population genetics
Richard Neher
Biozentrum, University of Basel
Viruses
 tobacco mosaic virus
(Thomas Splettstoesser, wikipedia)
tobacco mosaic virus
(Thomas Splettstoesser, wikipedia)
bacteria phage (adenosine, wikipedia)
influenza virus wikipedia
human immunodeficiency virus wikipedia
- rely on host to replicate
- little more than genome + capsid
- genomes typically 5-200k bases (+exceptions)
- most abundant organisms on earth $\sim 10^{31}$
Some viruses evolve a million times faster than animals
Animal haemoglobin
HIV protein

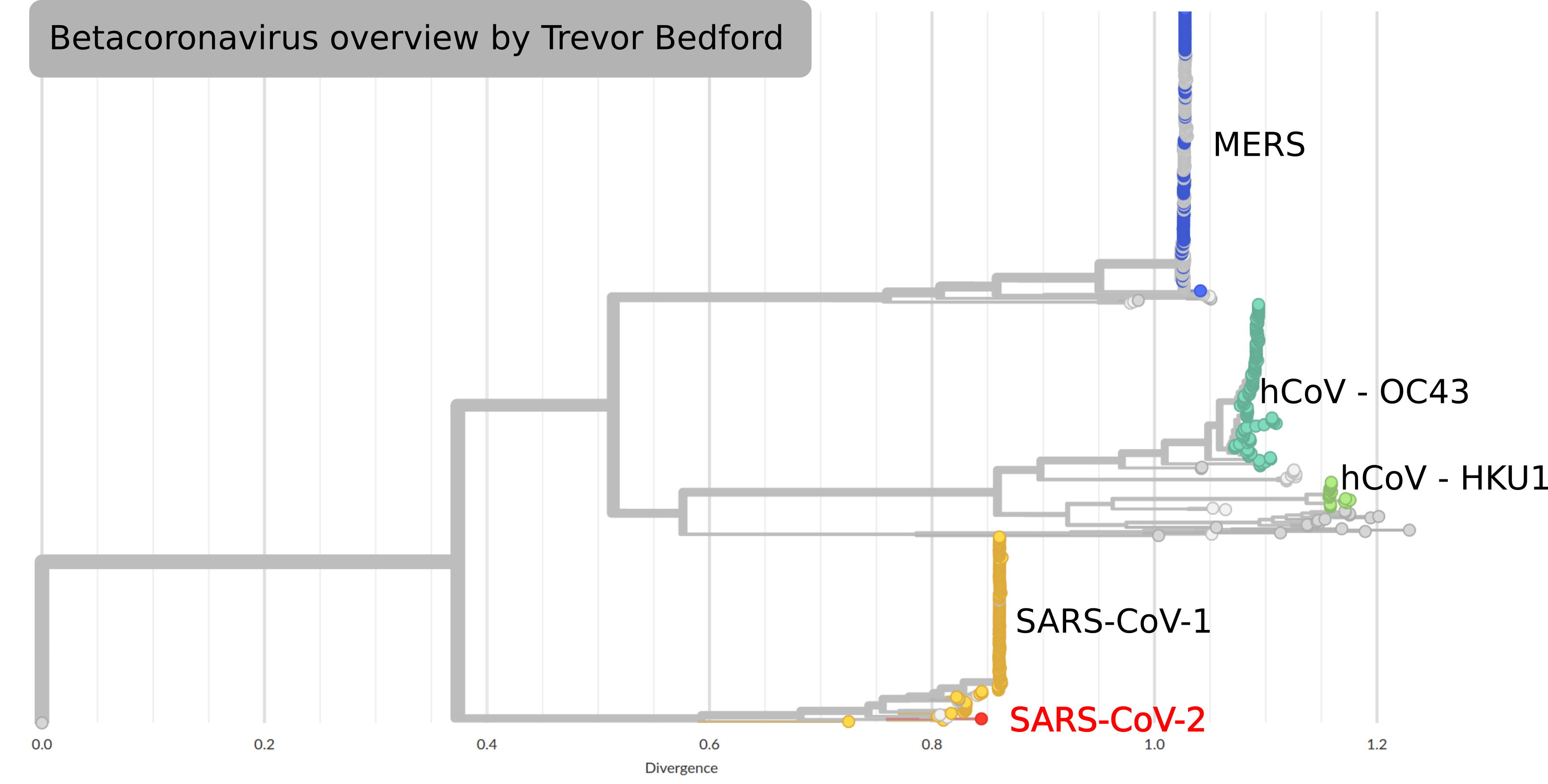 by Trevor Bedford
by Trevor Bedford
 by Trevor Bedford
by Trevor Bedford
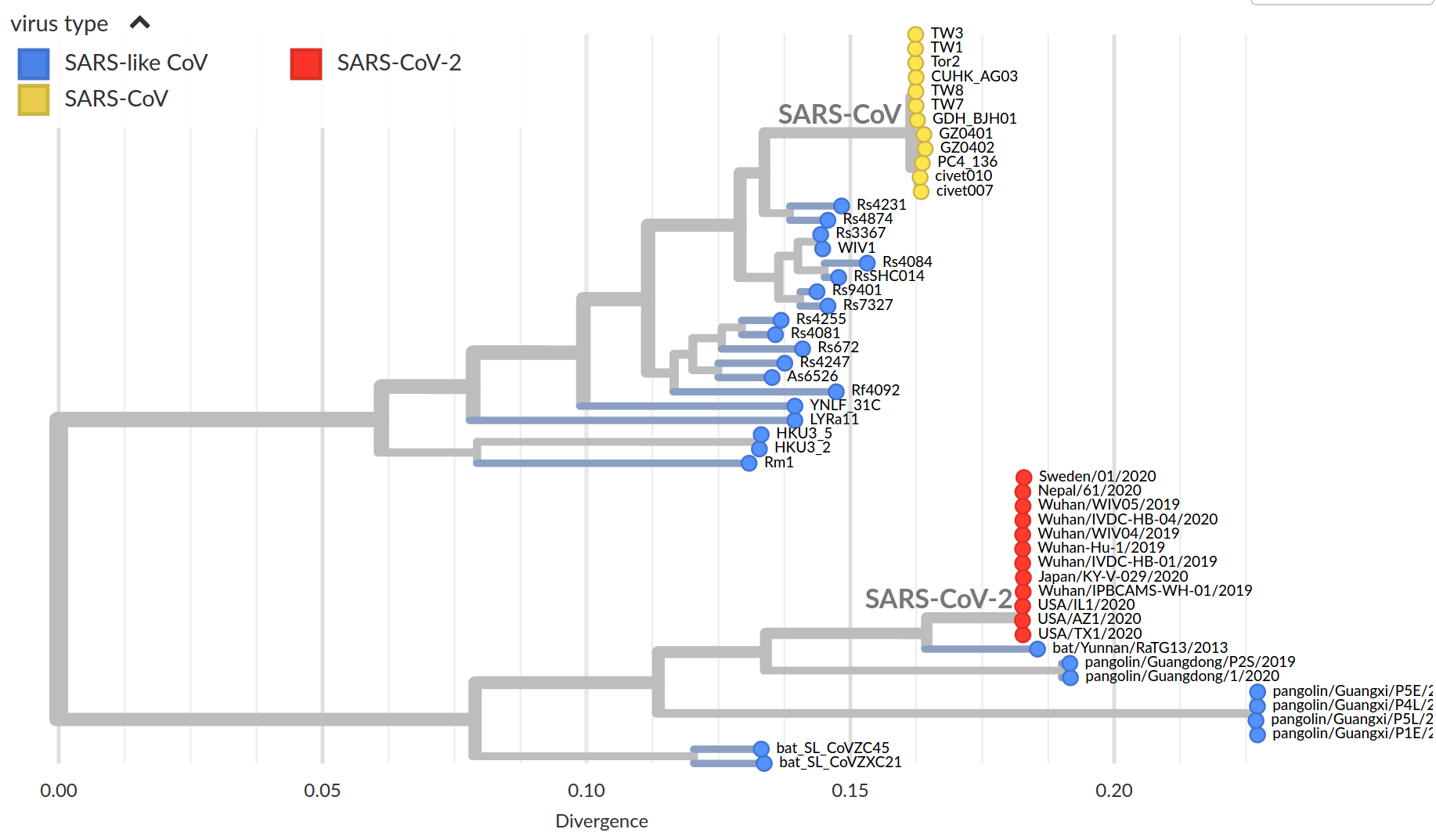
Tracking diversity and spread of SARS-CoV-2 in Nextstrain
Available data on Jan 26
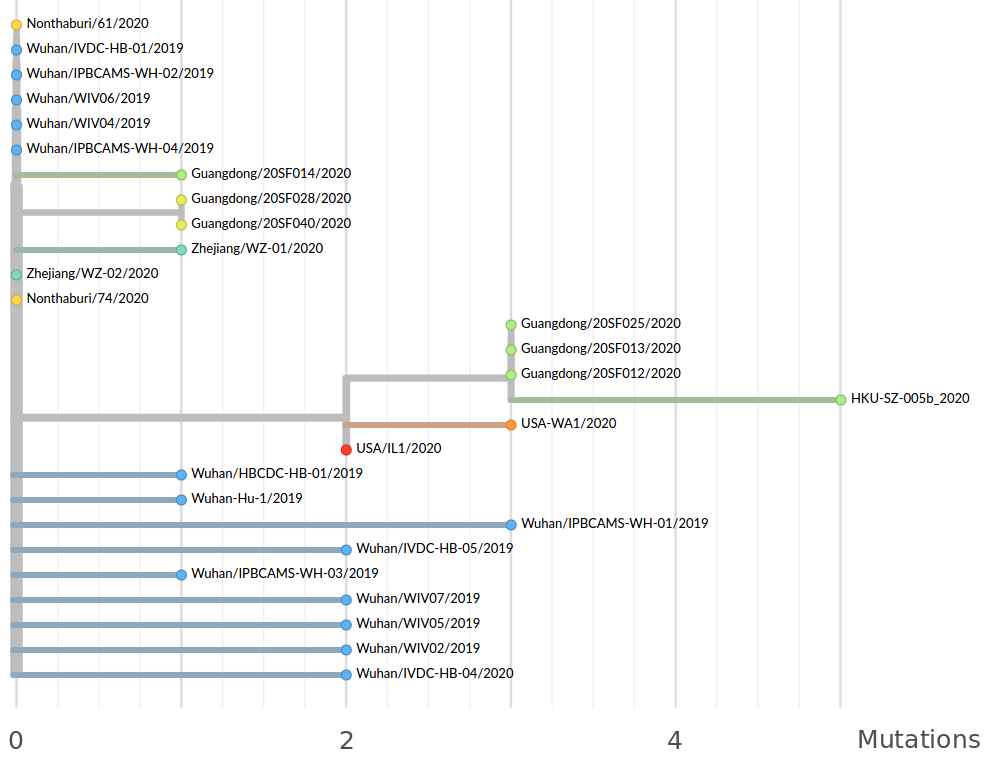
Early genomes differed by only a few mutations, suggesting very recent emergence
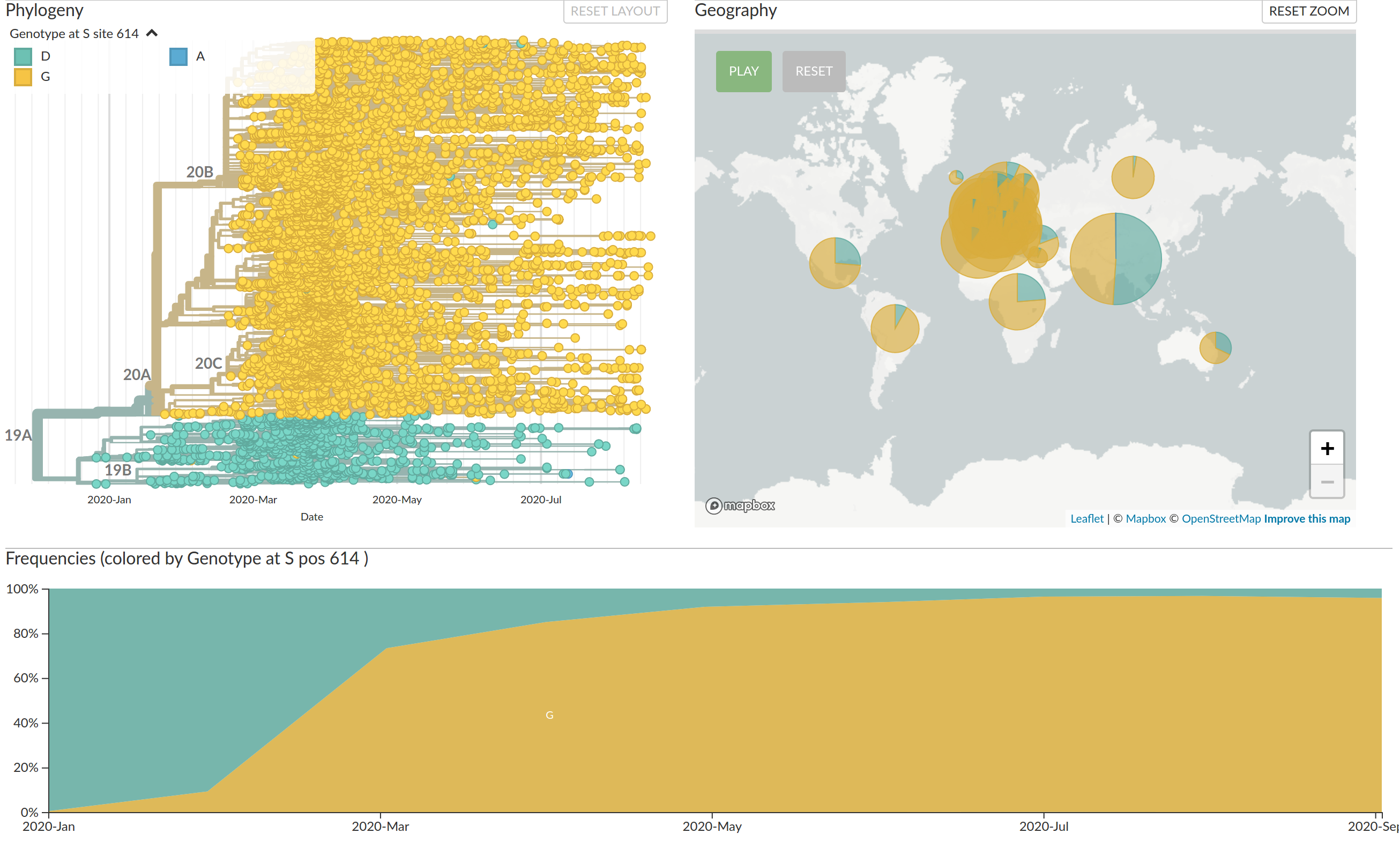
Interactive part on Nextstrain
Time-scaled phylogenies
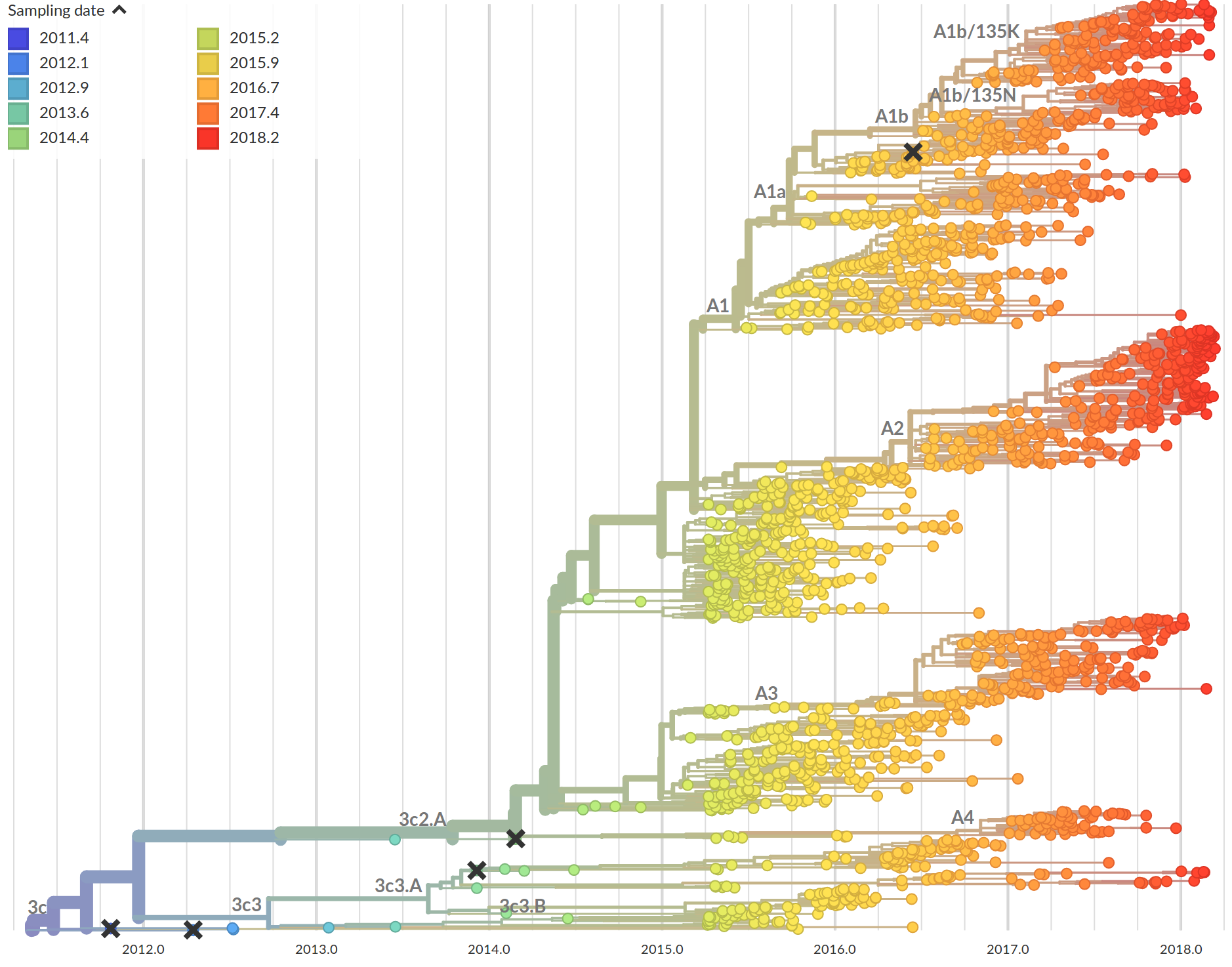
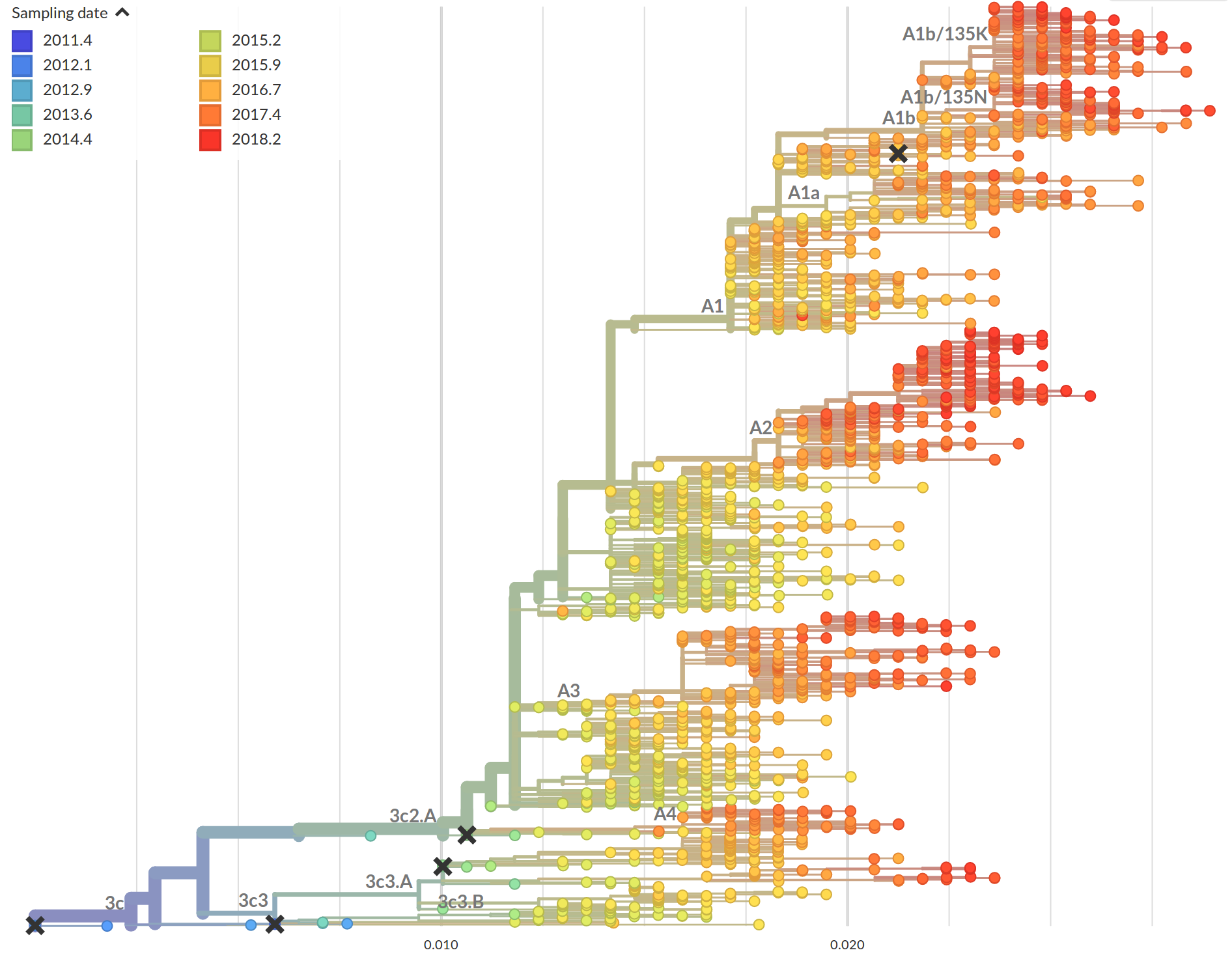
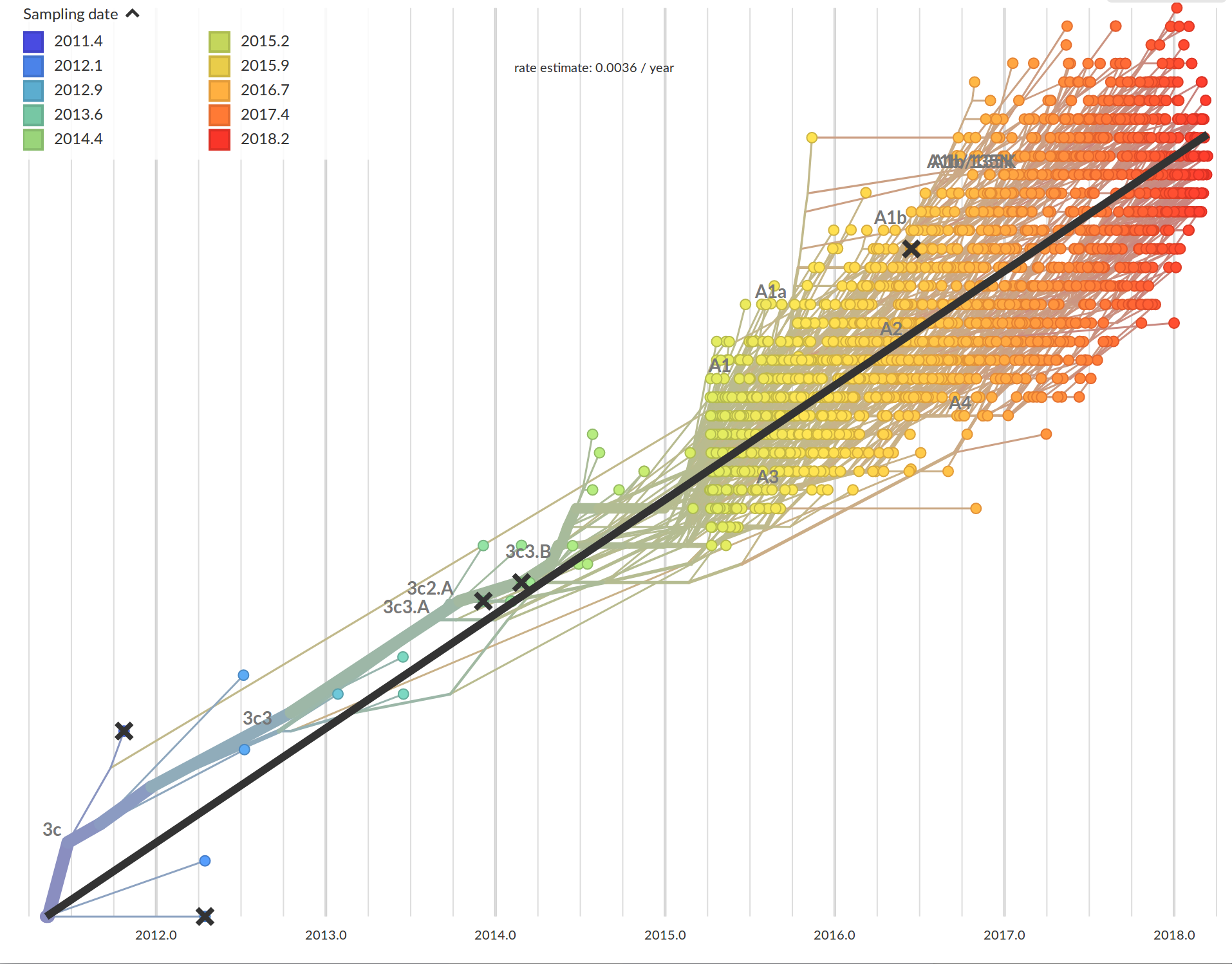
Tree building optimization with temporal constraints
- Time stamps single out a root
- Root can be found by optimizing root-to-tip regression
- BEAST: Markov-Chain Monte Carlo tree sampler
- If topology is correct, temporal constraints can be accounted for in linear time
- Multiple tools: treedate, LSD, treetime
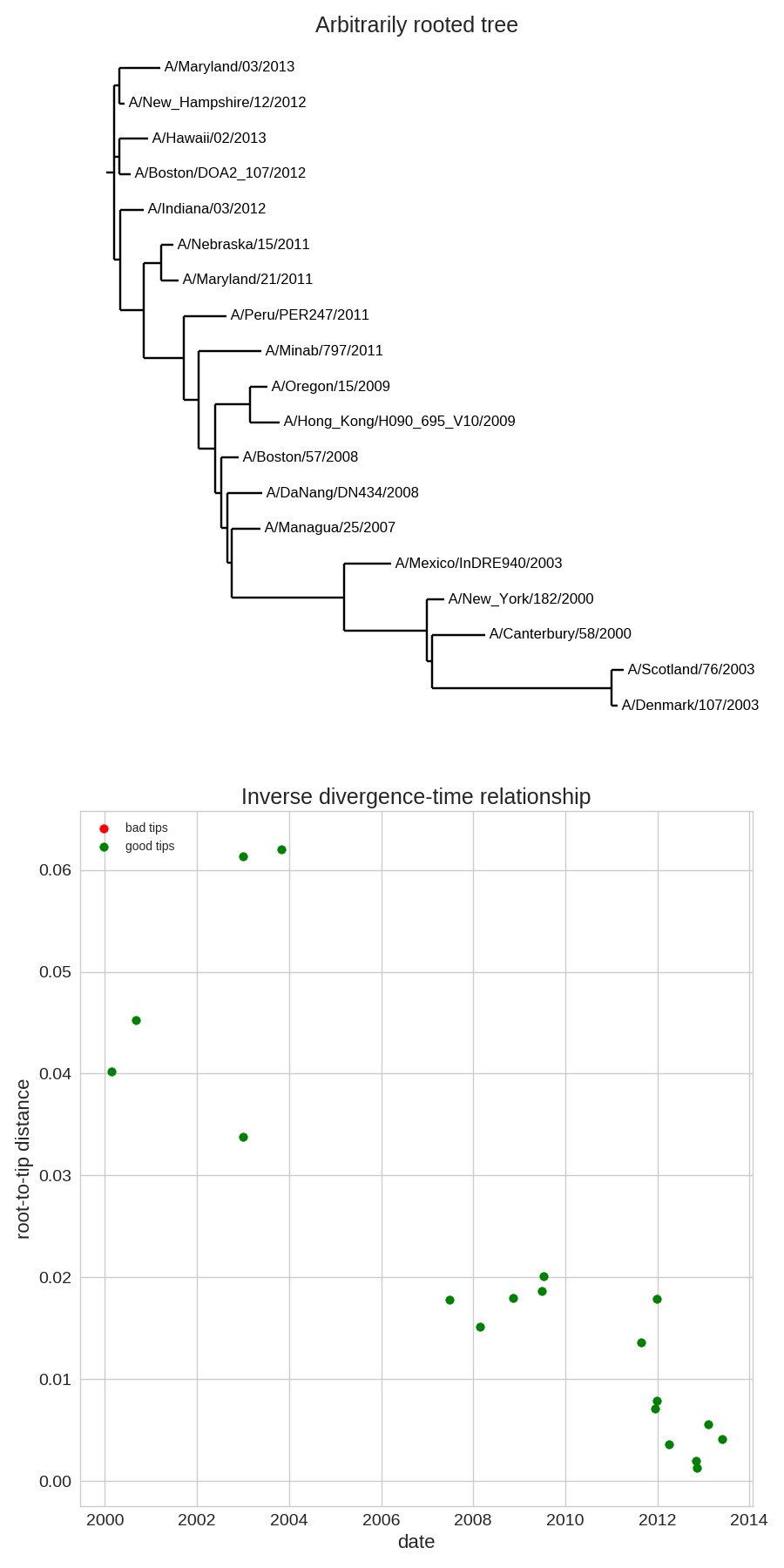
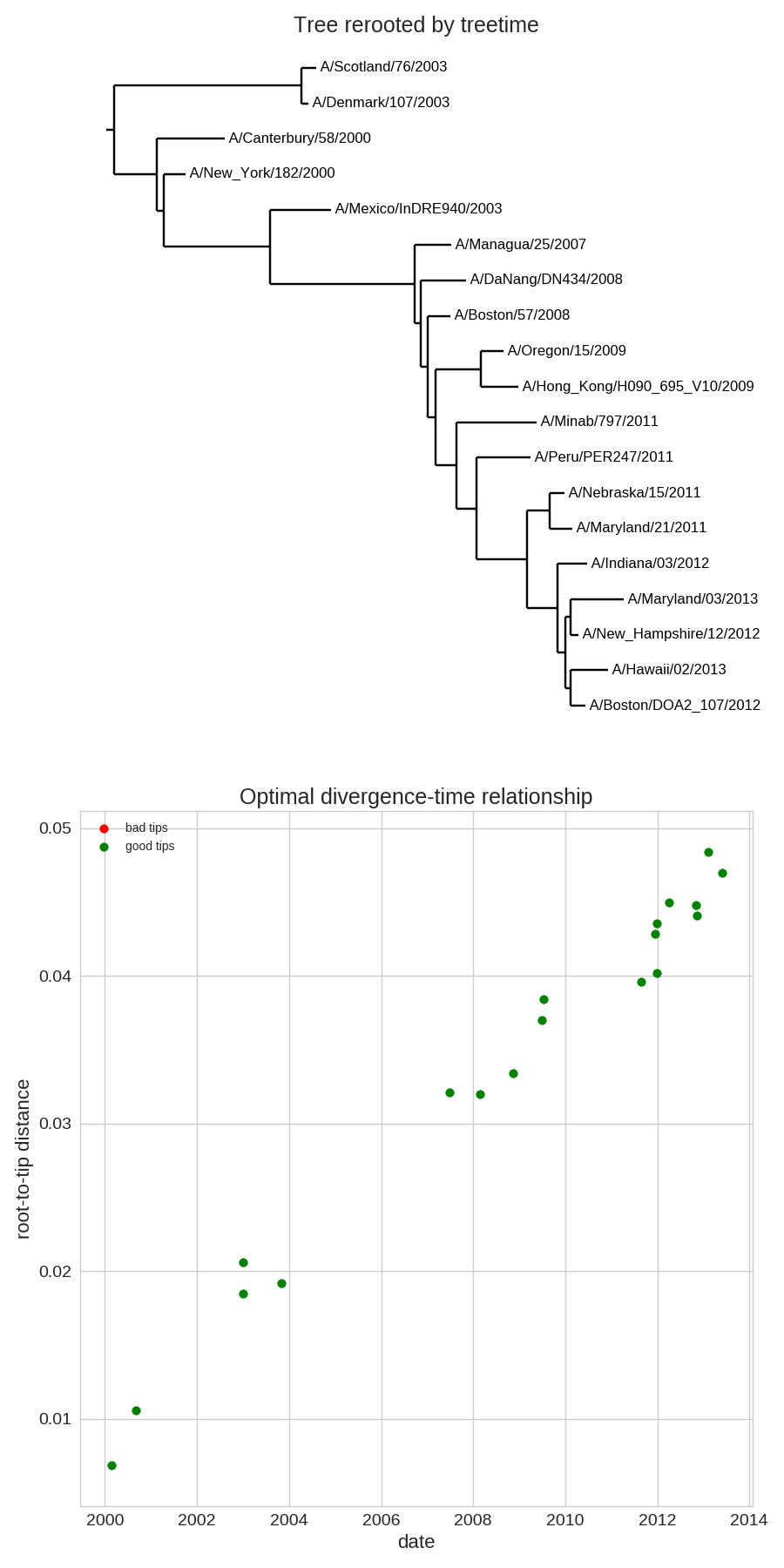
Time-scaled phylogenies

Attach sequences and dates
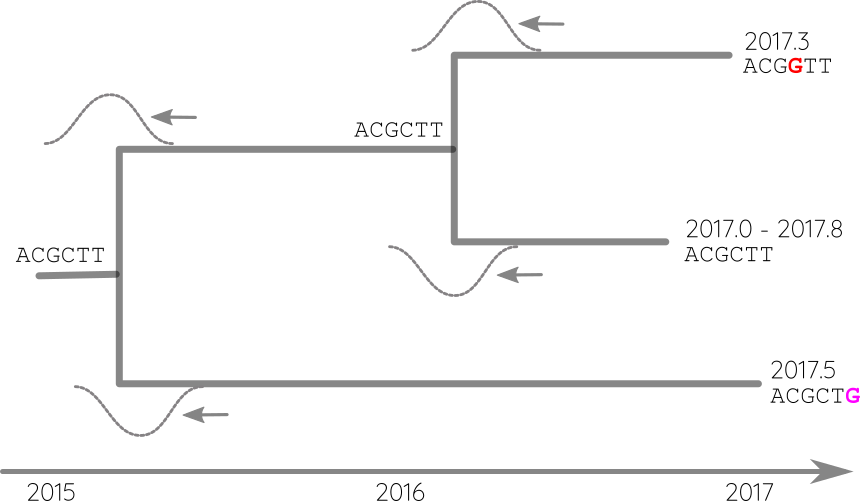
Propagate temporal constraints via convolutions
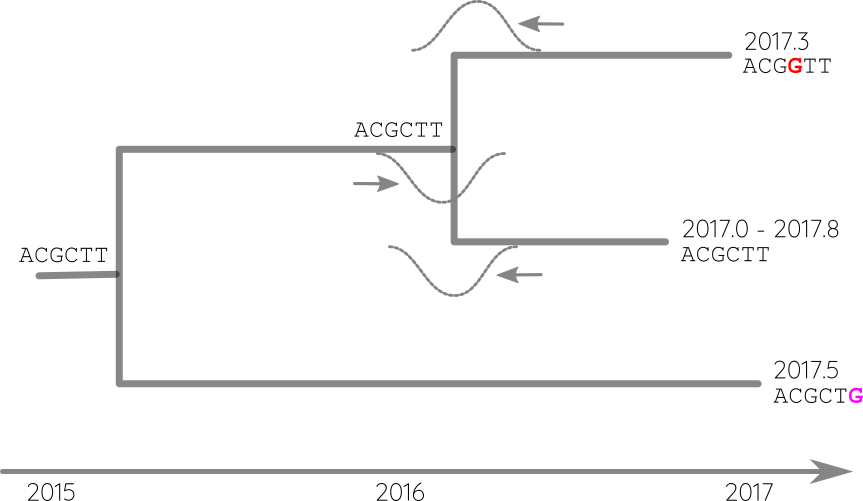
Integrate up-stream and down-stream constraints
Time-scaled phylogenies
- Calibration points can be longitudinal samples, ancient DNA or fossils
- Rates can vary between proteins and organisms from 0.01/year to $<10^{-8}$/y
- Some site change often, some rarely → saturation
- The apparent rate changes over time
- Divergence times are often under estimated.
treetime.ch
- Download these SARS-CoV-2 sequences
- go to clades.nextstrain.org
- upload the sequence data above (will process for a few minutes)