Tracking and interpreting novel SARS-CoV-2 variants
Richard Neher
Biozentrum & SIB, University of Basel
slides at neherlab.org/202106_DPG.html
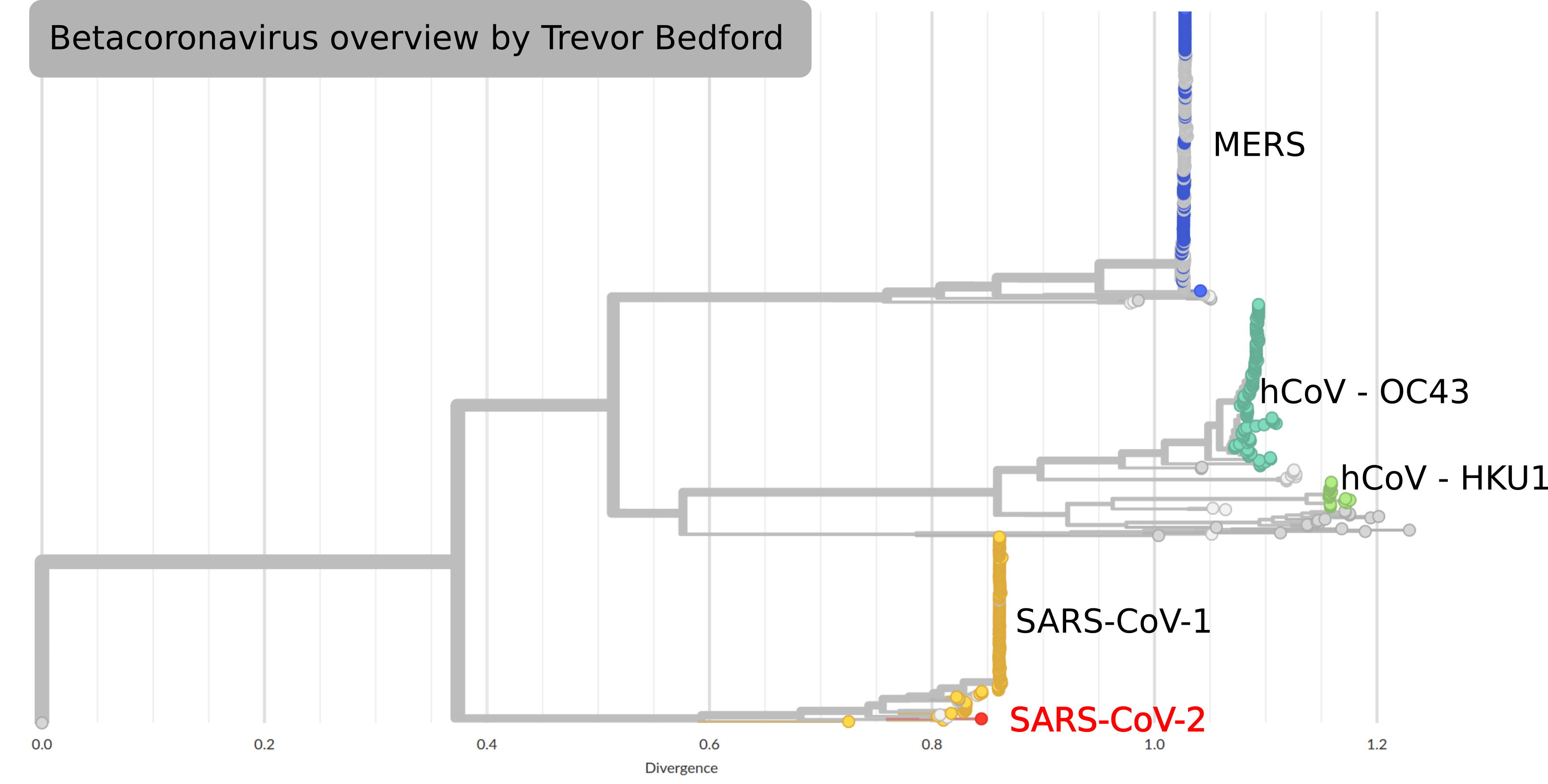 by Trevor Bedford
by Trevor Bedford
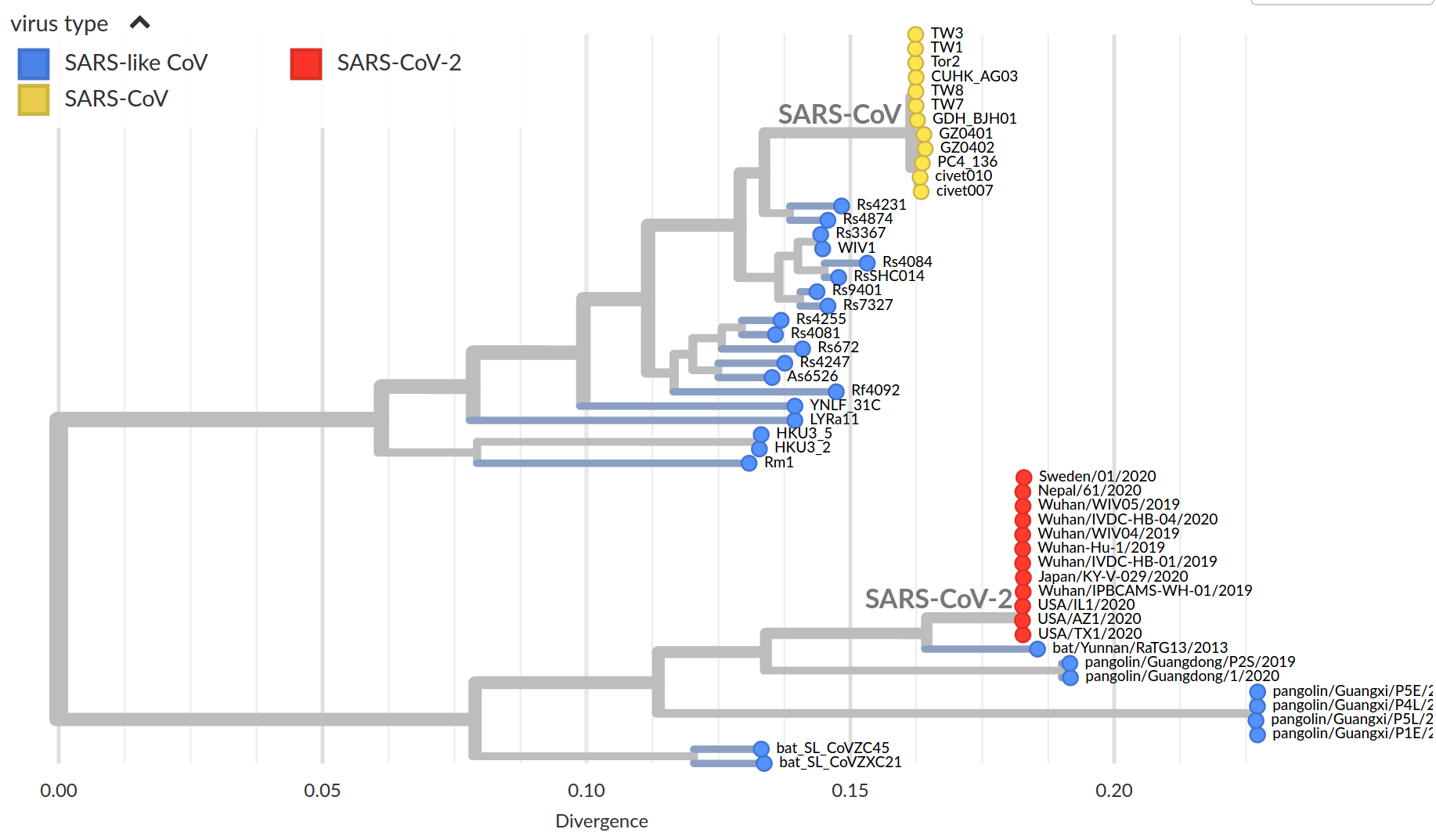 by Trevor Bedford
by Trevor Bedford
Available data on Jan 26 2020
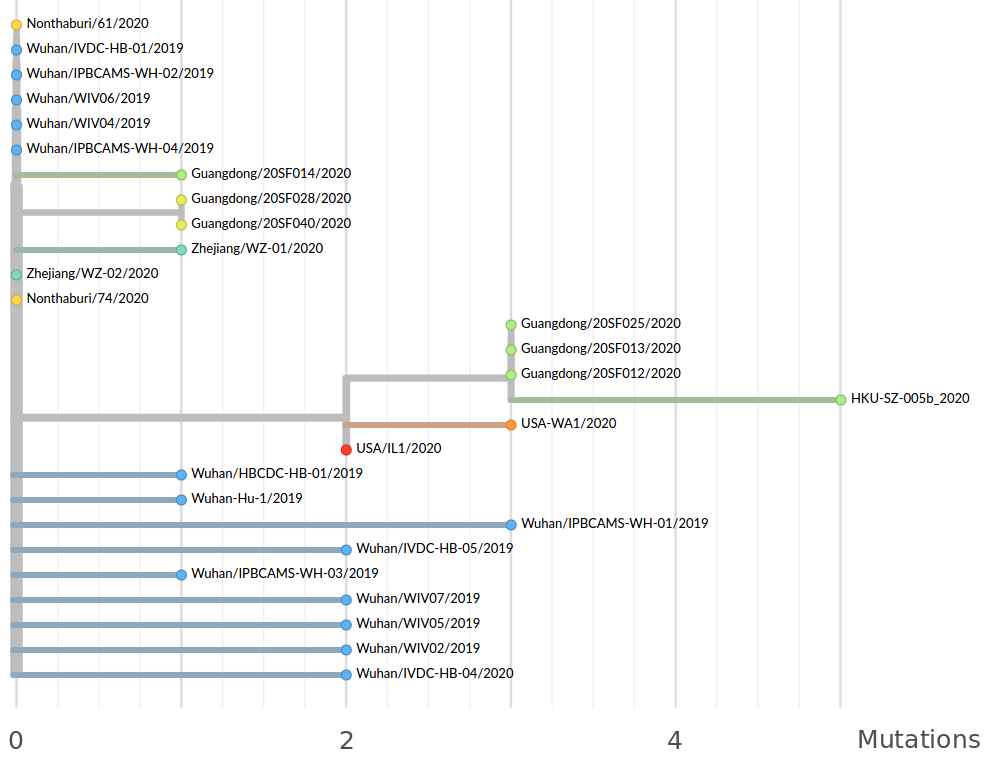
Early genomes differed by only a few mutations, suggesting very recent emergence
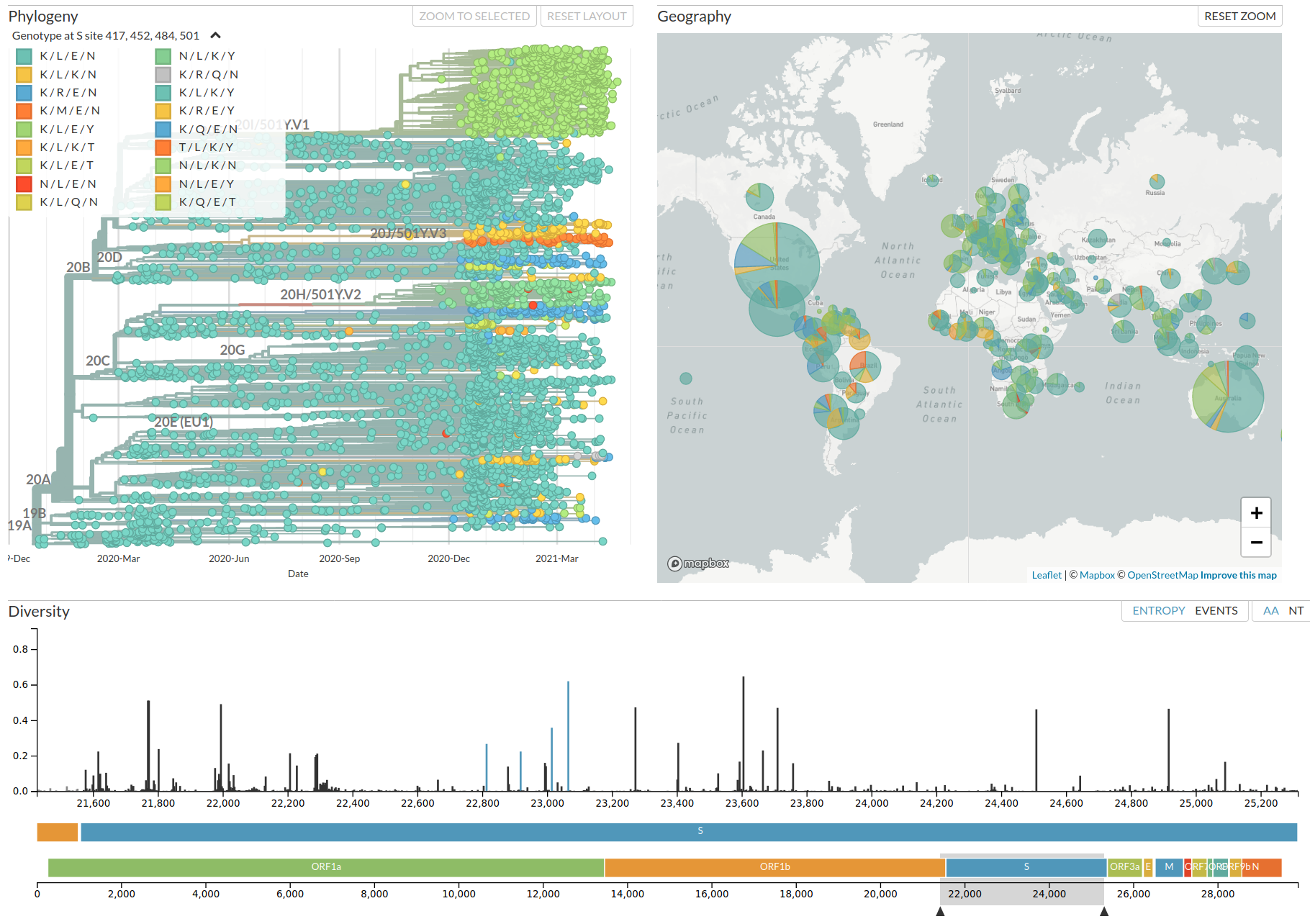
nextstrain.org
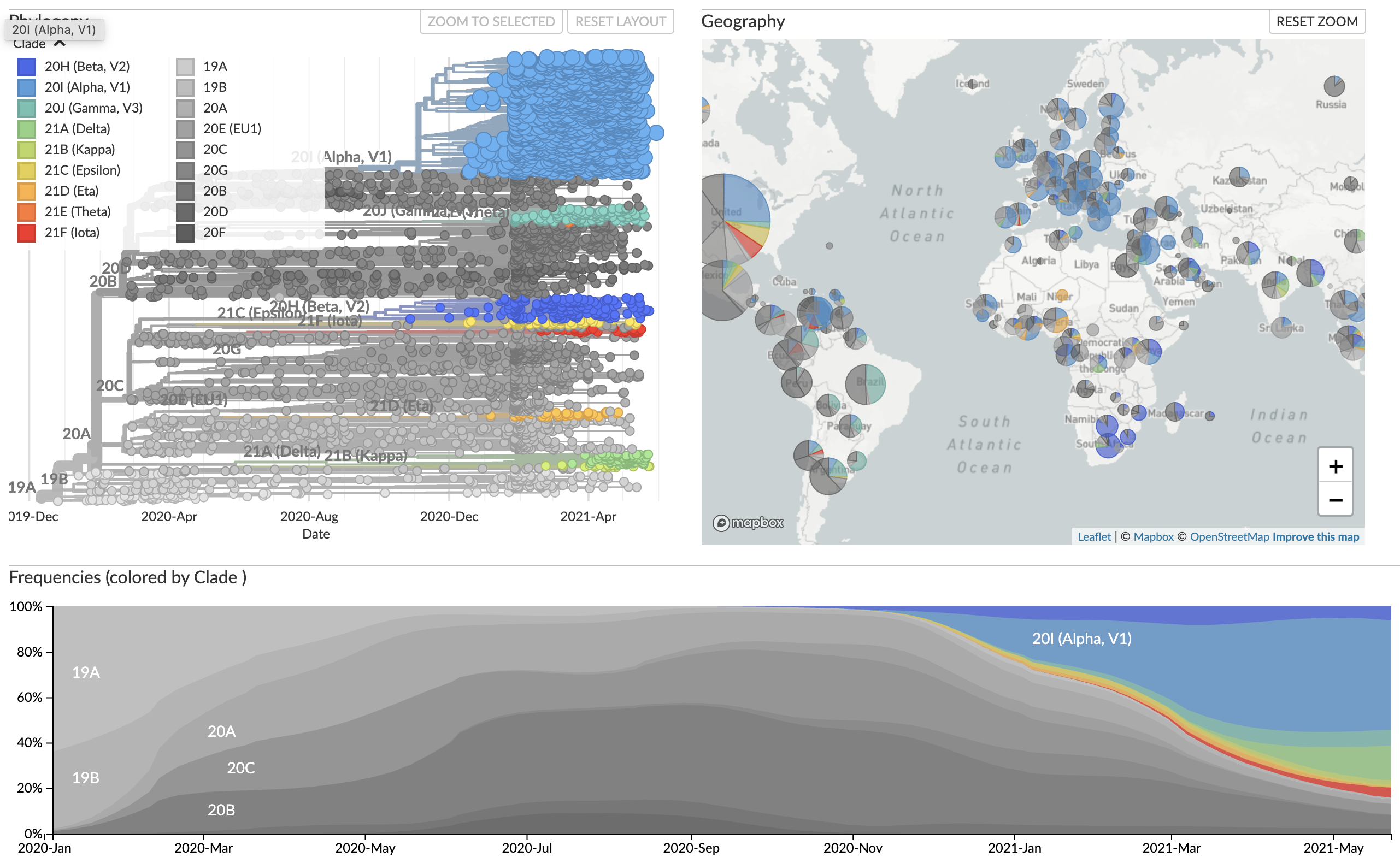
VoCs have more mutations than expected...
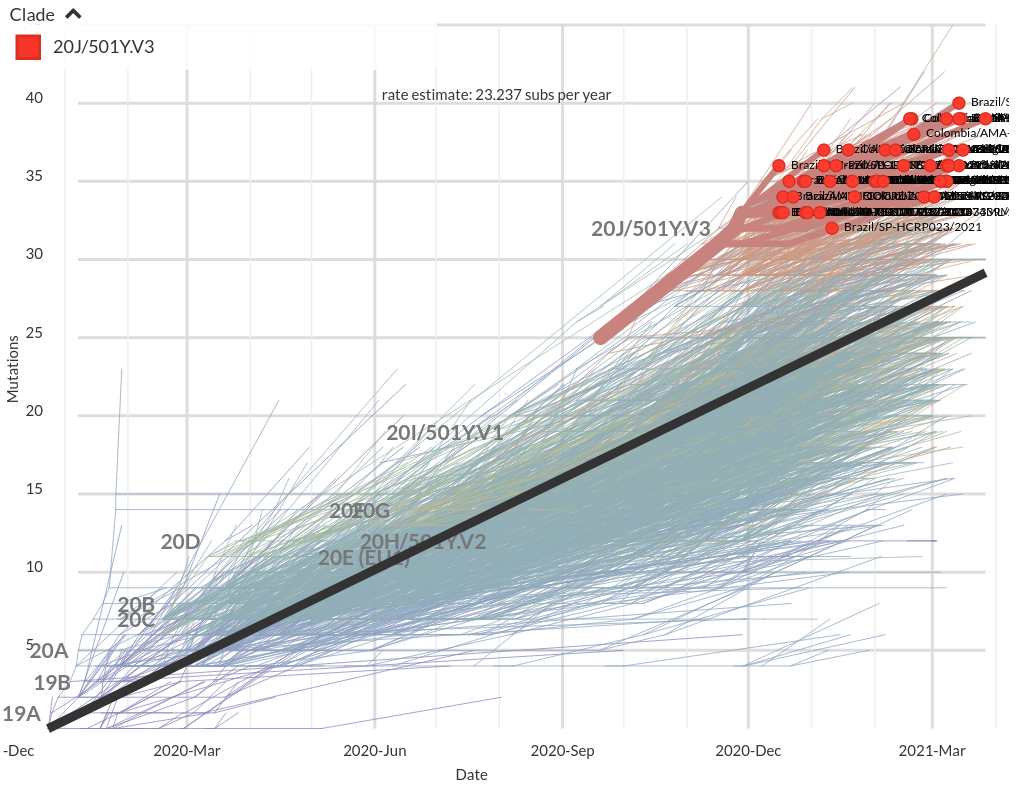 nextstrain
nextstrain
VoCs and VoIs: rapid converging evolution
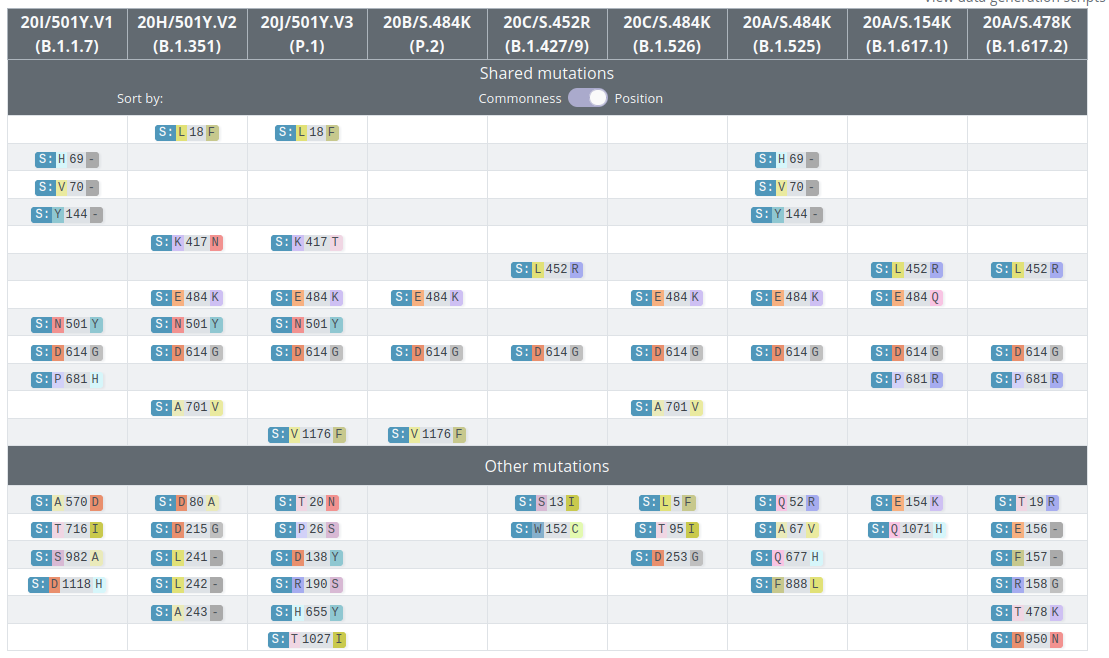 covariants.org by Emma Hodcroft
covariants.org by Emma Hodcroft
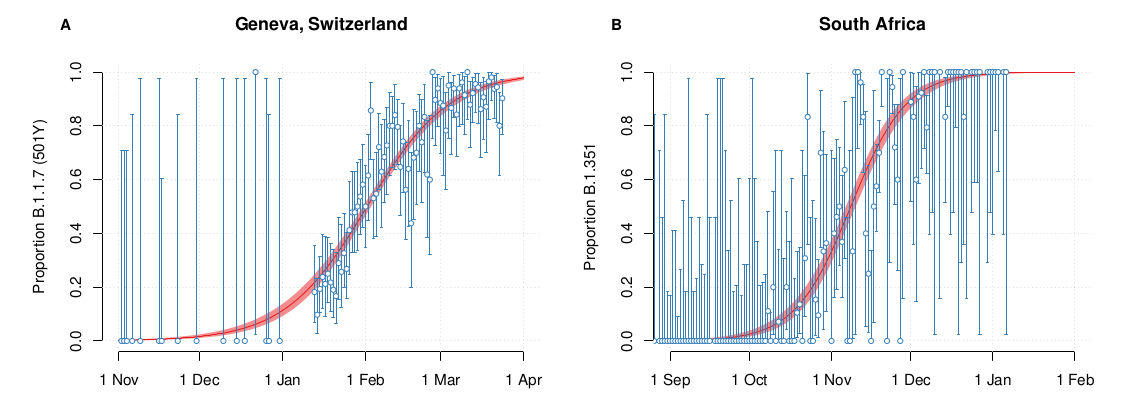
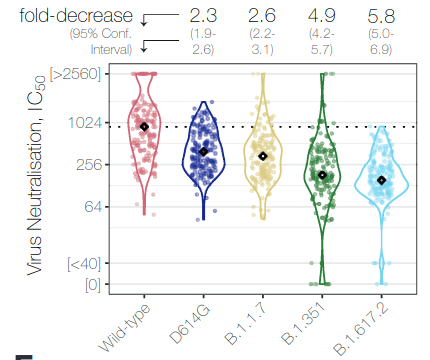
VoCs partially escape neutralizing antibodies and transmit faster
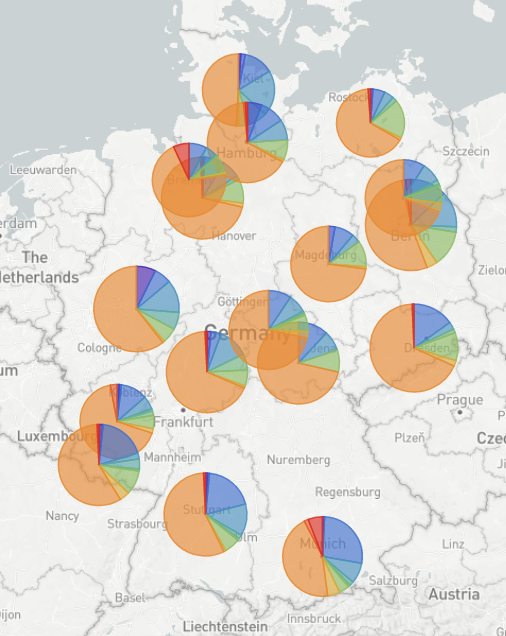
Nextstrain builds for different countries
nextstrain.org/groups/neherlab
- Started as part of the surveillance effort in Switzerland
- Most European countries -- updated twice a week
- Heavy focus on sequences from the last 4 weeks (but still subsampled in many cases)
- Administrative divisions for some countries
- Variant specific builds via CoVariants
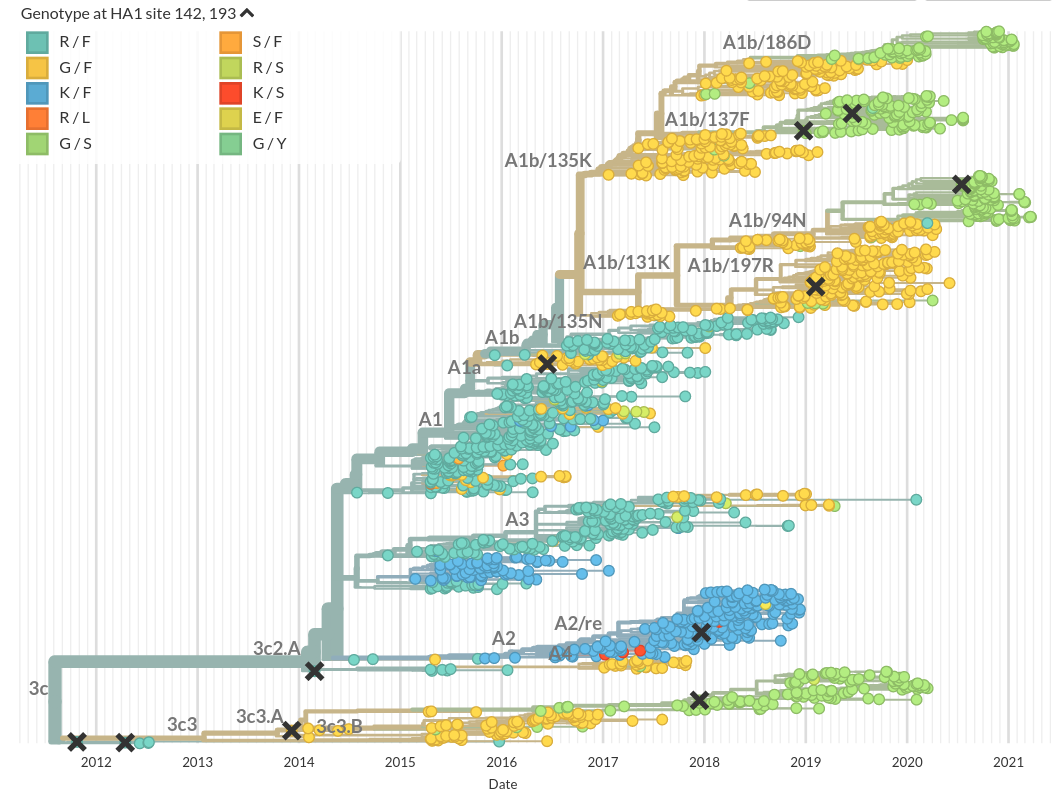
Similarities to influenza virus evolution
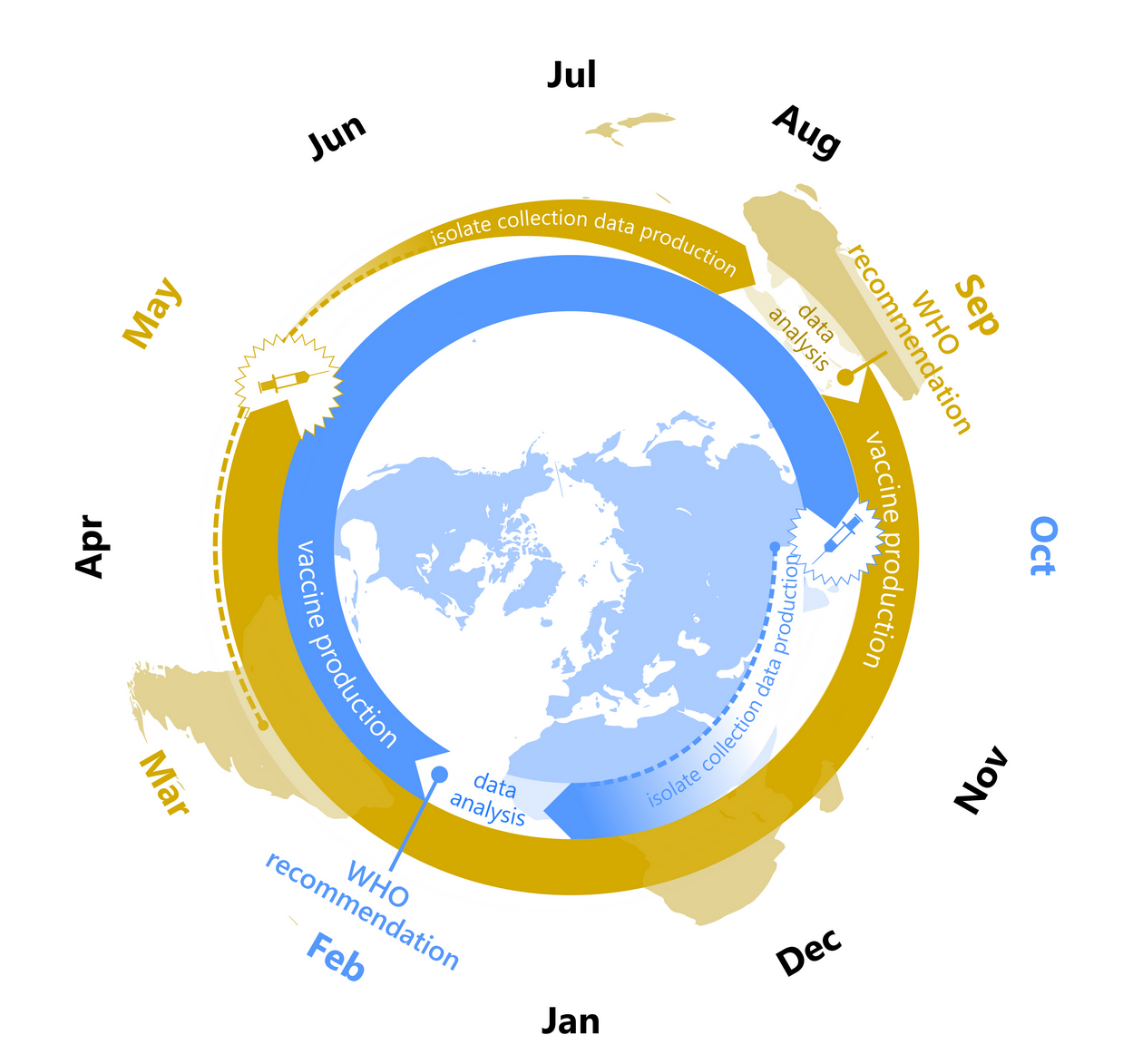 Klingen et al, Trends in MicroBio, 2018
Klingen et al, Trends in MicroBio, 2018
Acknowledgments
Trevor Bedford and his lab -- terrific collaboration since 2014

especially James Hadfield, Emma Hodcroft, Ivan Aksamentov, Moira Zuber, and Tom Sibley
Allie Greaney and Jesse Bloom for discussion of the dms data
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID

