Patterns of early diversity of SARS-CoV-2
Richard Neher
Biozentrum & SIB, University of Basel
slides at neherlab.org/202106_ParisGroup.html
Do early patterns of diversity give us clues about the origin of SARS-CoV-2?
Maybe, but interpreting these patterns is hard an much uncertainty remains.
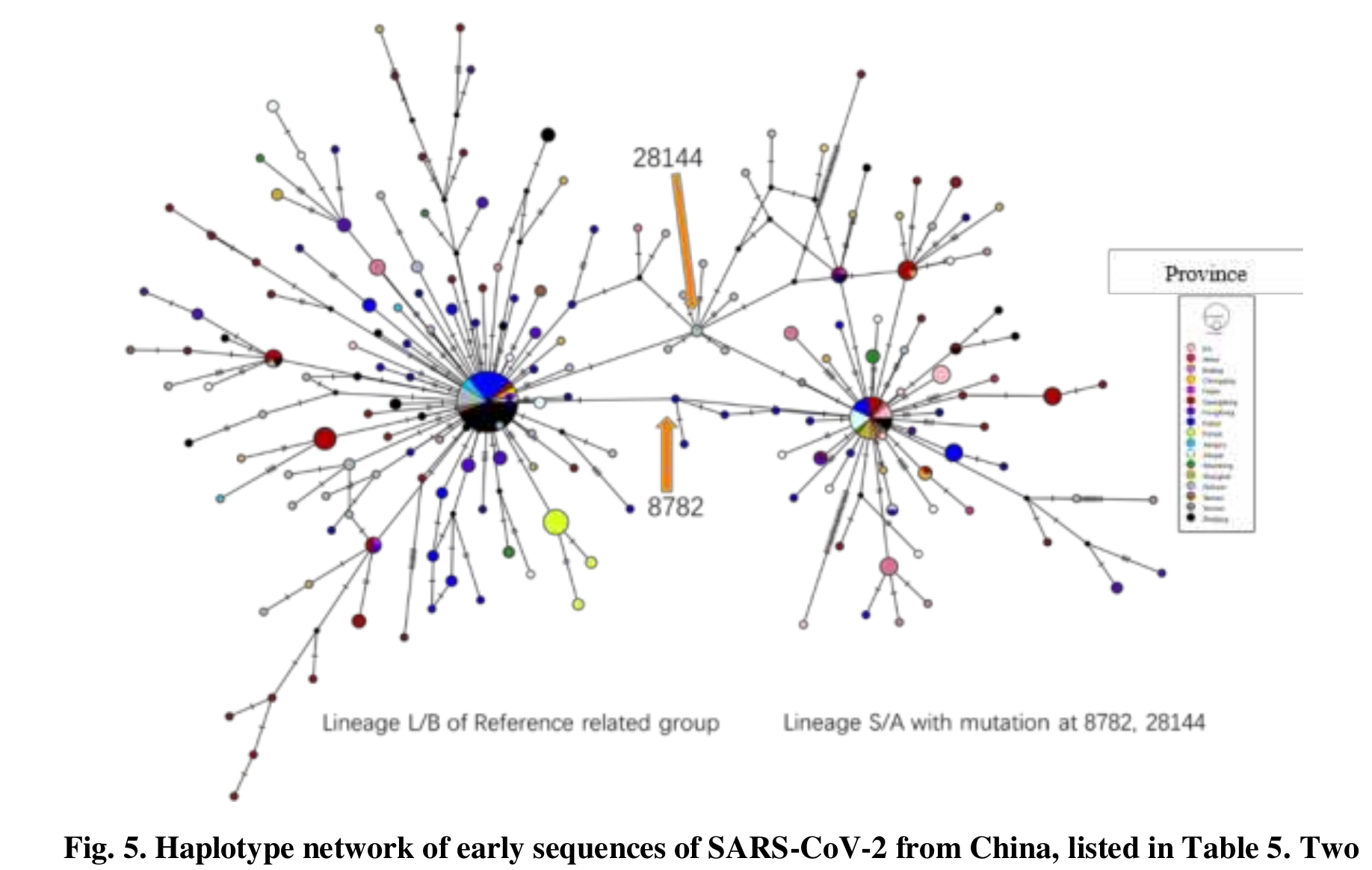 WHO report
WHO report
Multi-market hypothesis

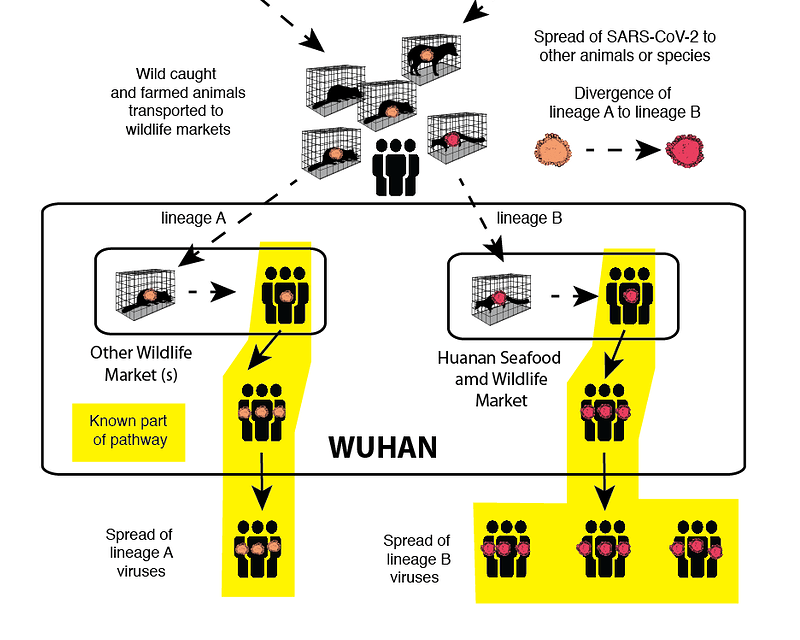 Garry, virological.org, 2021
Garry, virological.org, 2021
Plausible roots of current SC2 diversity
- 19A/lineage B: Huanan market cluster, favored by models without explicit outgroups, implies 2 reversions
- 19B/lineage A: 2 mutations closer to bat viruses, would result in an earlier TMRCA
- Guangdong cluster: 3 mutations closer to bat viruses, epidemiologically less plausible, still earlier TMRCA
- Other viruses close to bat viruses: mostly one-offs, some reversions are expected by chance
Root inferred by Pekar et al
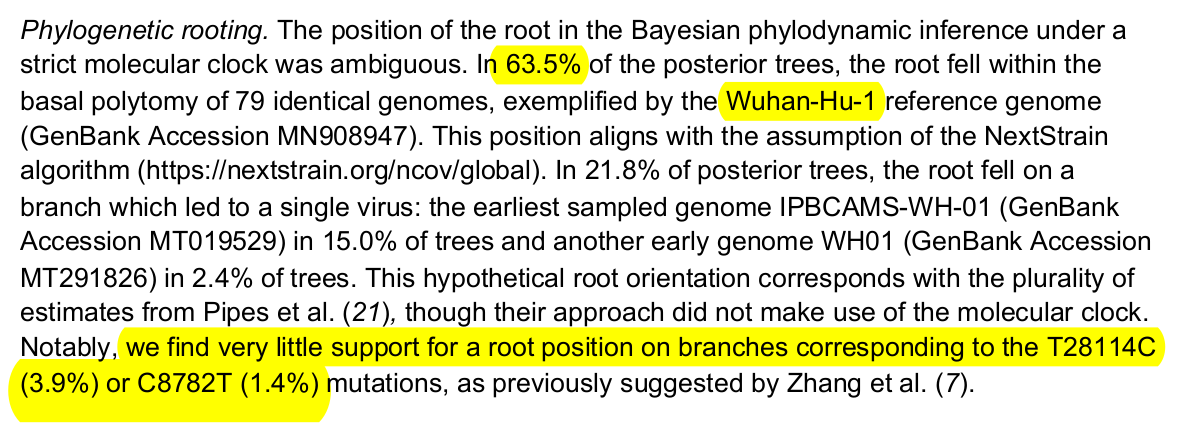 Pekar et al
Pekar et al
Similarity to bat viruses
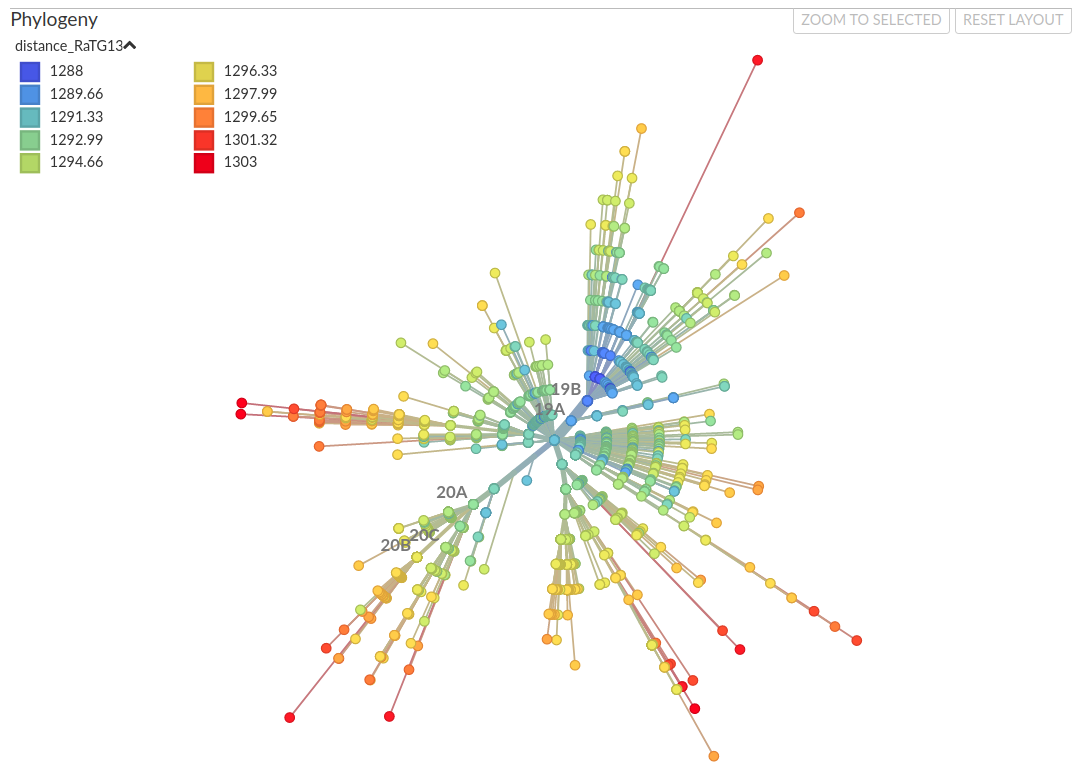
The 8782/28144 split
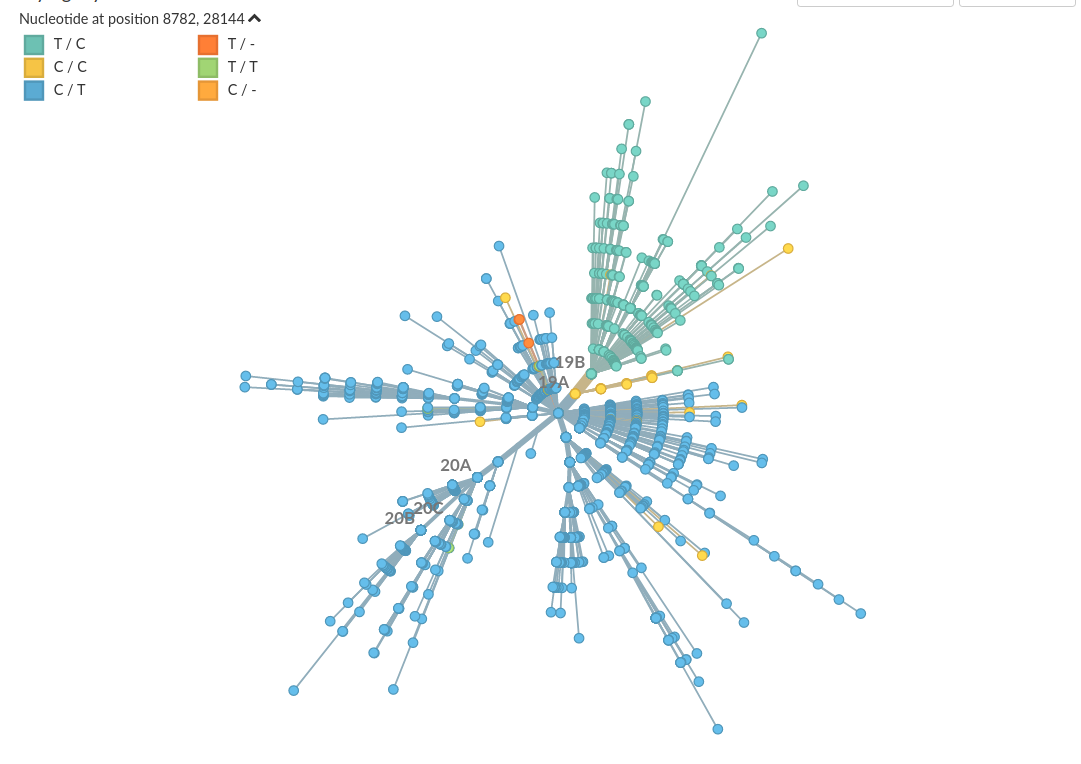
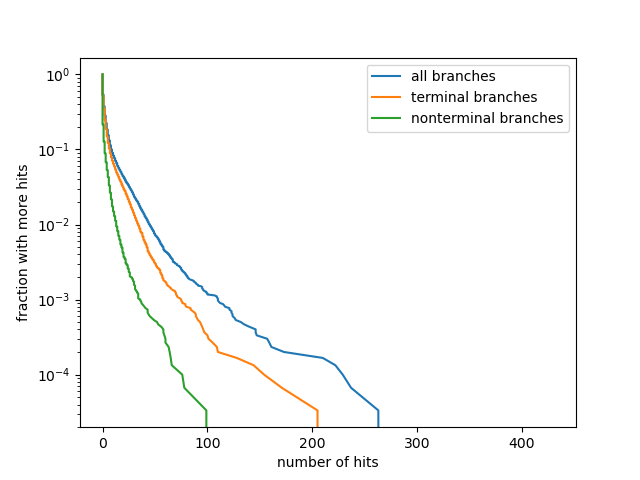
Homoplasies and Reversions to bat-like states
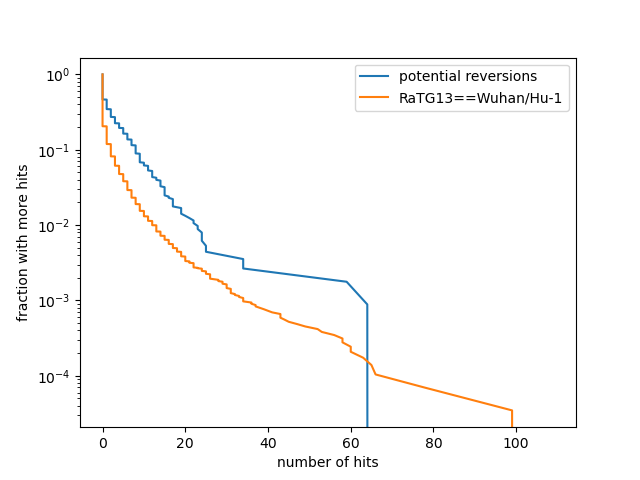
Homoplasies and Reversions to bat-like states (non-terminals only)
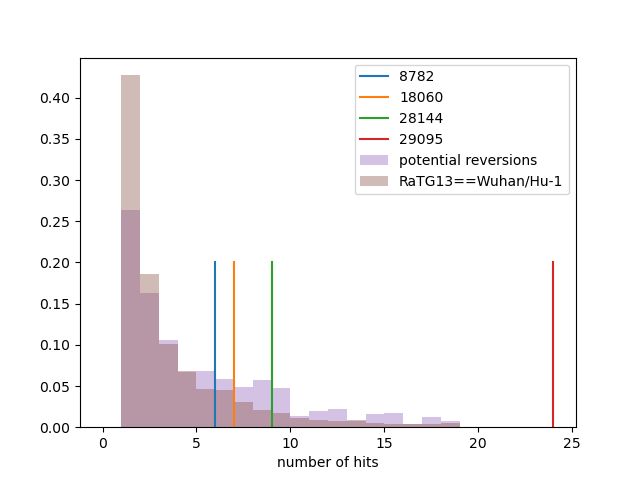
Conclusions
- Both 19A/19B rootings are plausible
- Early epidemiological dynamics is expected to be very stochastic, sampling focussed on 19A (lineage B, Huanan cluster)
→ epi-evidence weak in my opinion - Positions 8782 and 21844 are reasonably stable
→ points towards 19B (lineage A) as root - Some genomes need to be interpreted with caution
Acknowledgments
Trevor Bedford and his lab -- terrific collaboration since 2014

especially James Hadfield, Emma Hodcroft, Ivan Aksamentov, Moira Zuber, John Huddleston, and Tom Sibley
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID

