Adaptation and convergent evolution of SARS-CoV-2 and other human RNA viruses
Richard Neher
Biozentrum & SIB, University of Basel
slides at neherlab.org/202109_GfV.html
Available data on Jan 26 2020
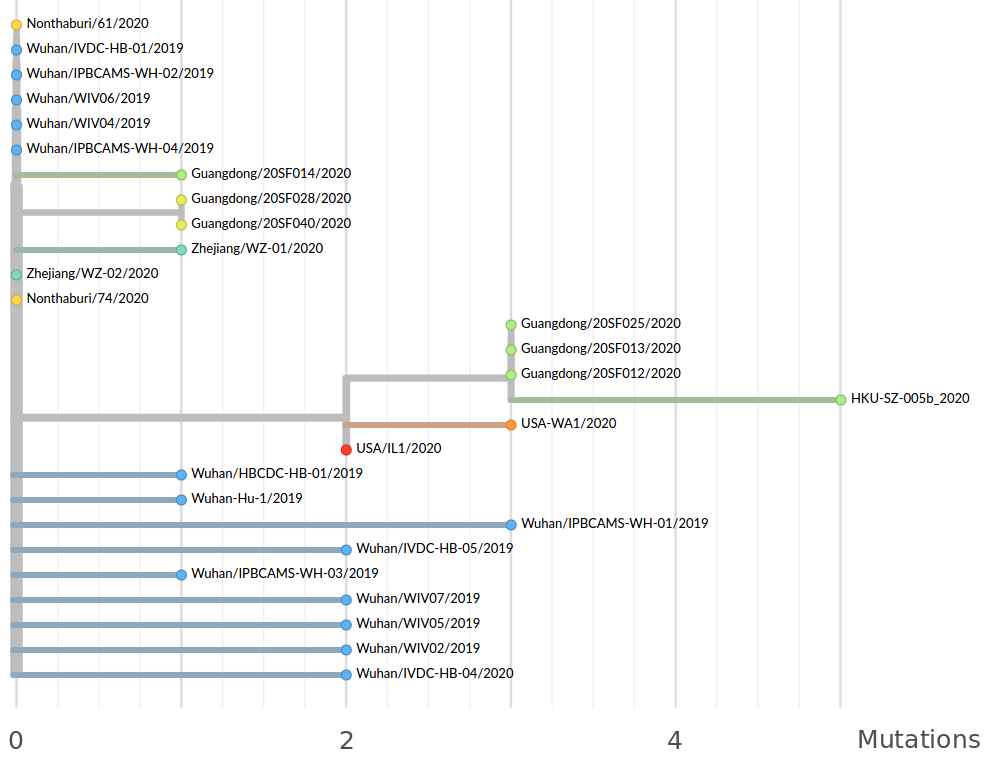
Early genomes differed by only a few mutations, suggesting very recent emergence
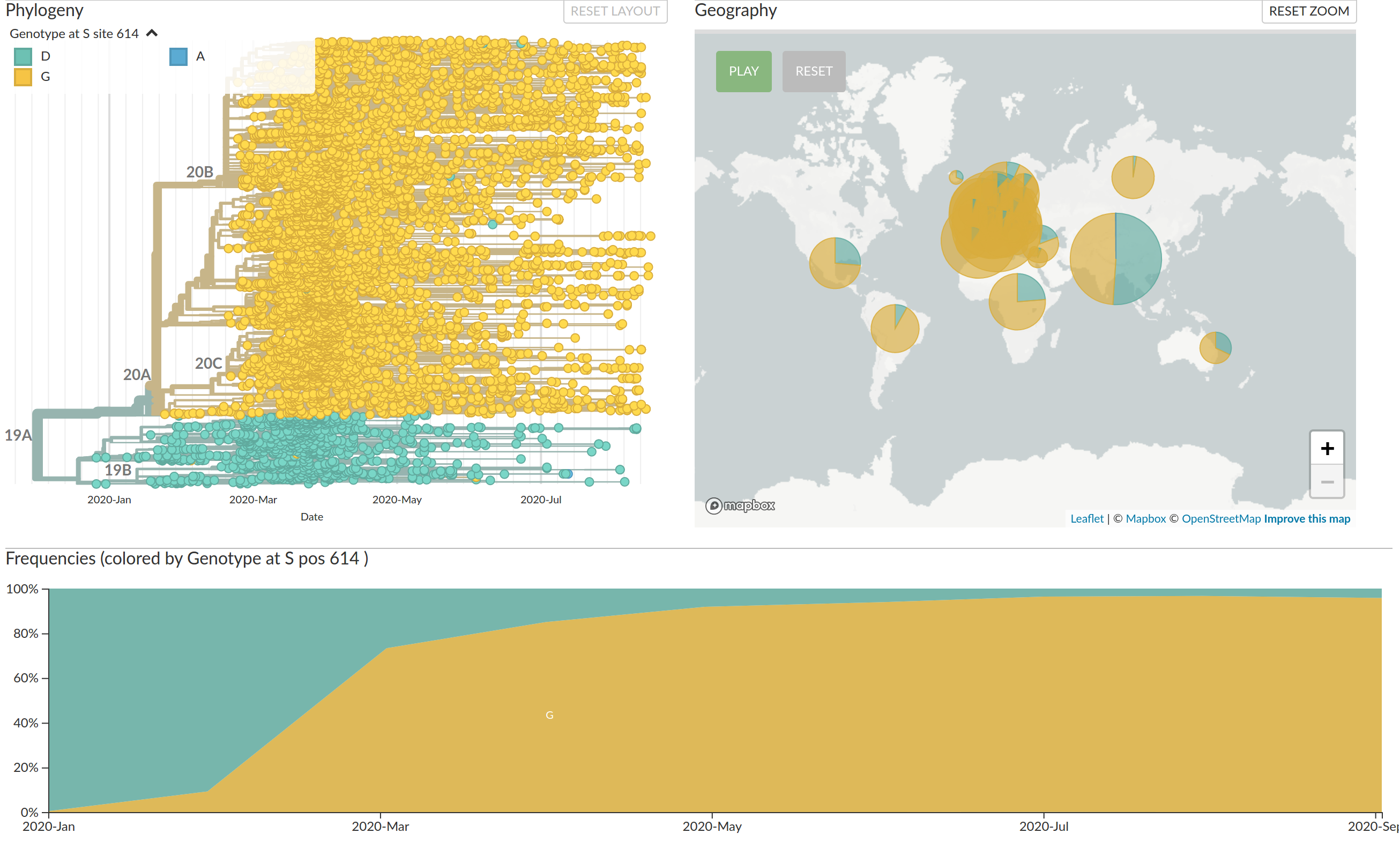
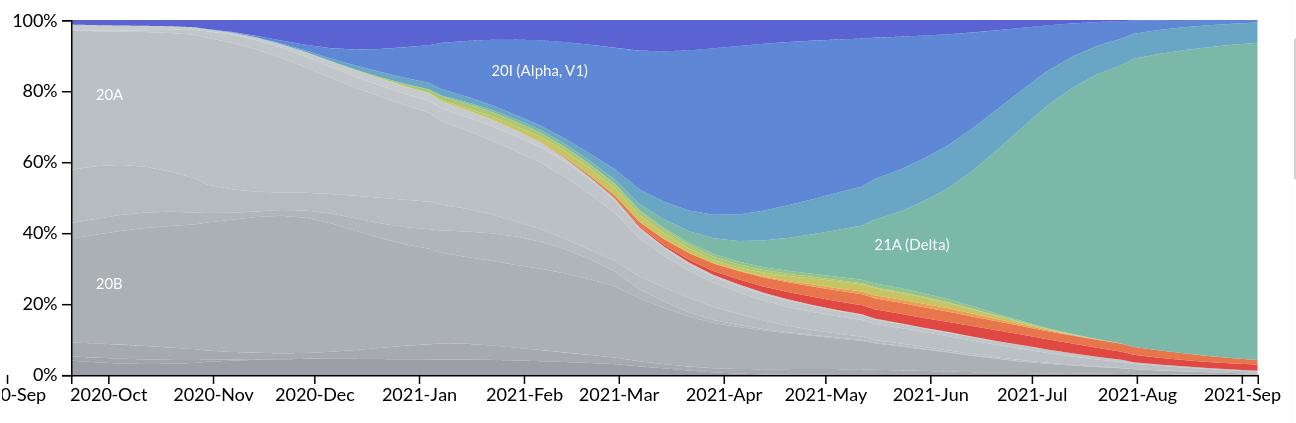
Emergence and dominance of VoCs
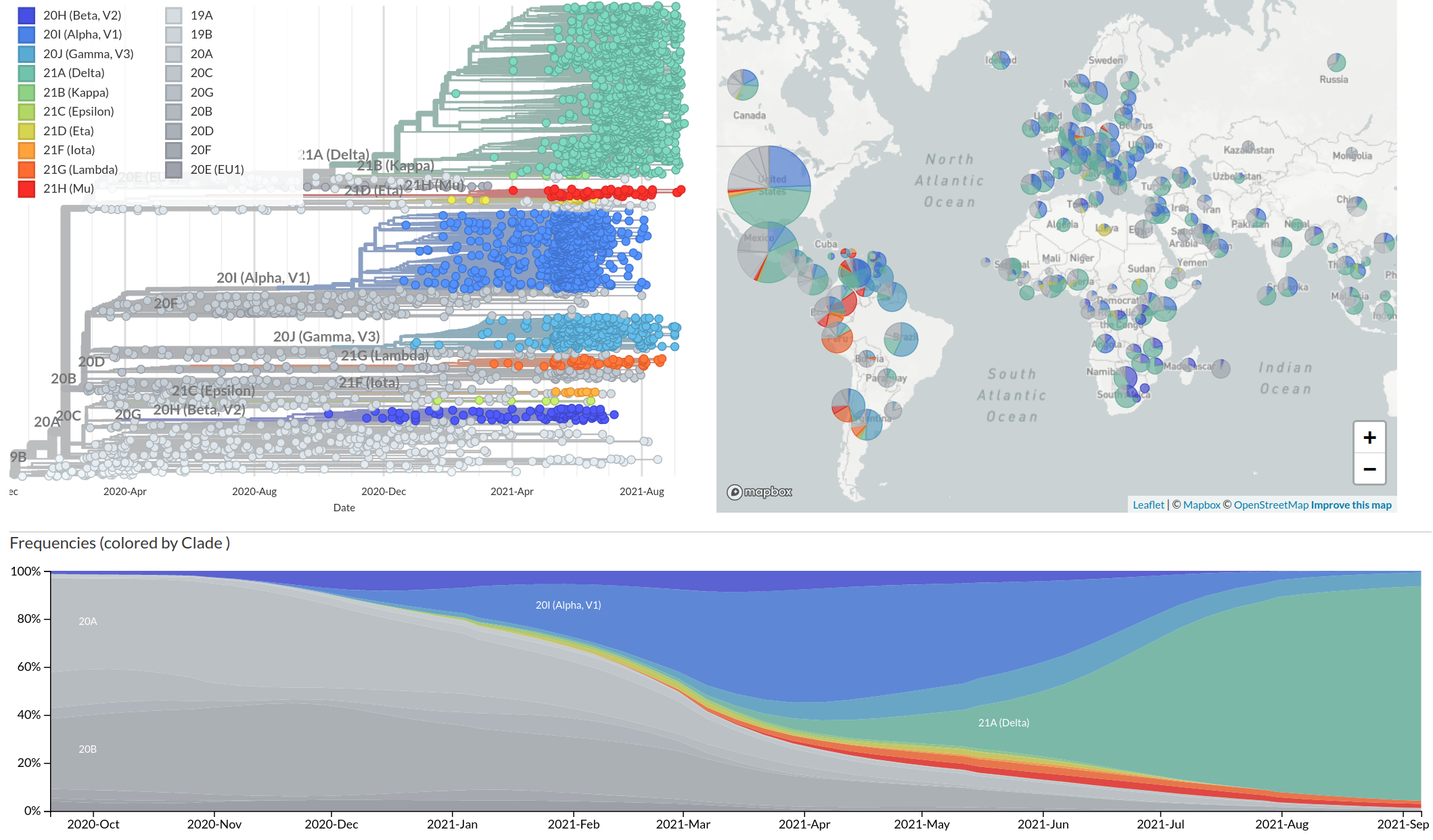
Real-time tracking on Nextstrain
VoCs have more mutations than expected...
 nextstrain
nextstrain
VoCs and VoIs: rapid converging evolution
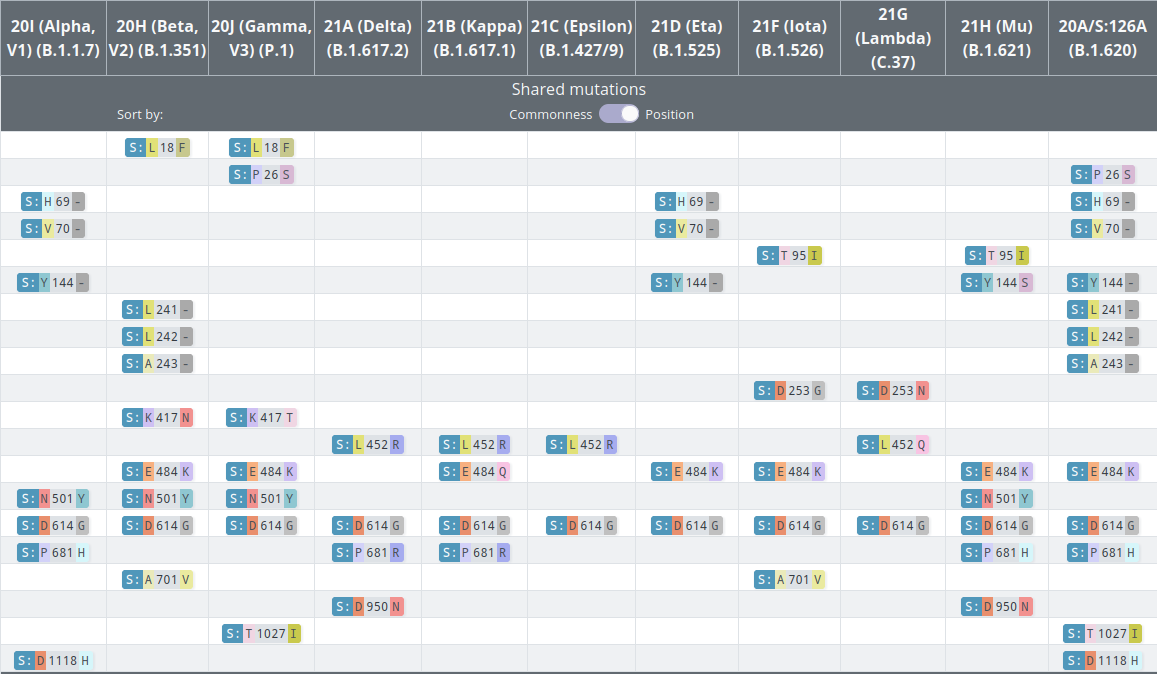 covariants.org by Emma Hodcroft
covariants.org by Emma Hodcroft
VoCs have been replacing other strains quickly
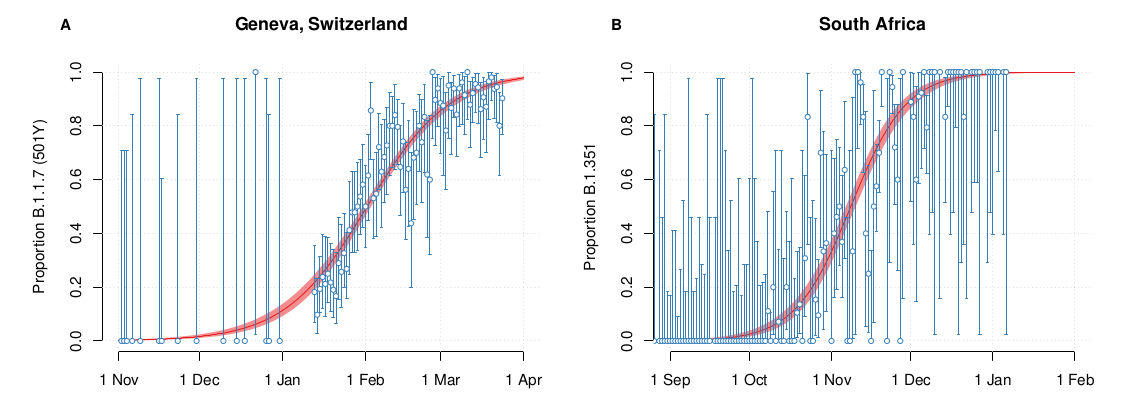
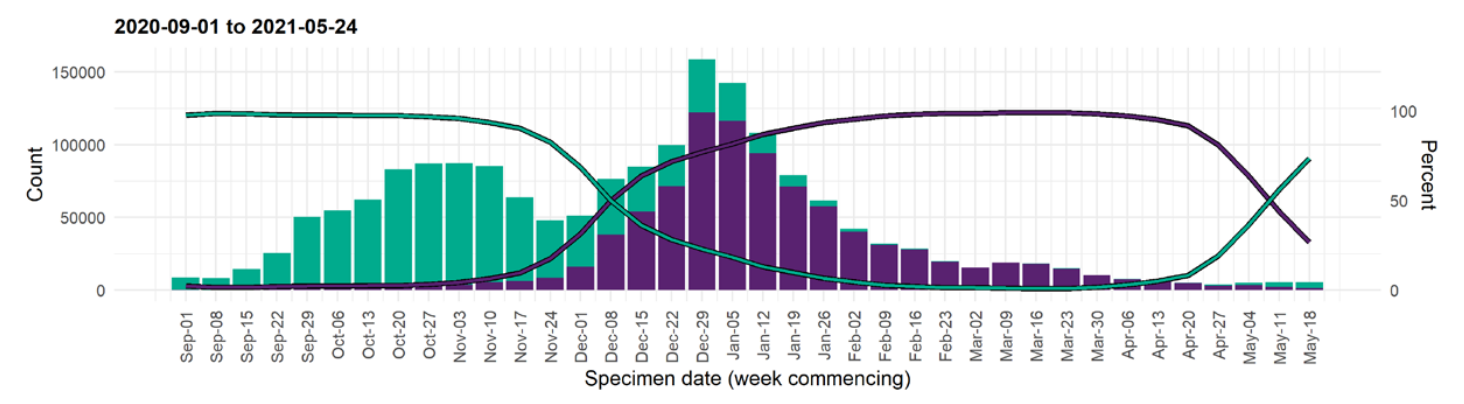 S-gene positivity in the UK
S-gene positivity in the UK
SARS-CoV-2 variants can become dominant without transmission advantage
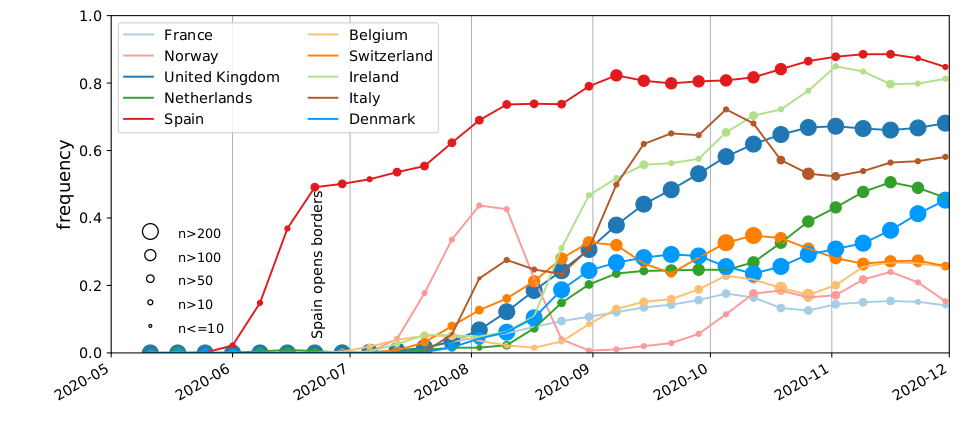
- Large differences in incidence (Spain vs most of Europe in Summer 2020)
- High travel volume during summer 2020
- Demographics with more social contacts
How do we tell early on whether a variant is concerning?
How unusual are the patterns of convergent evolution?
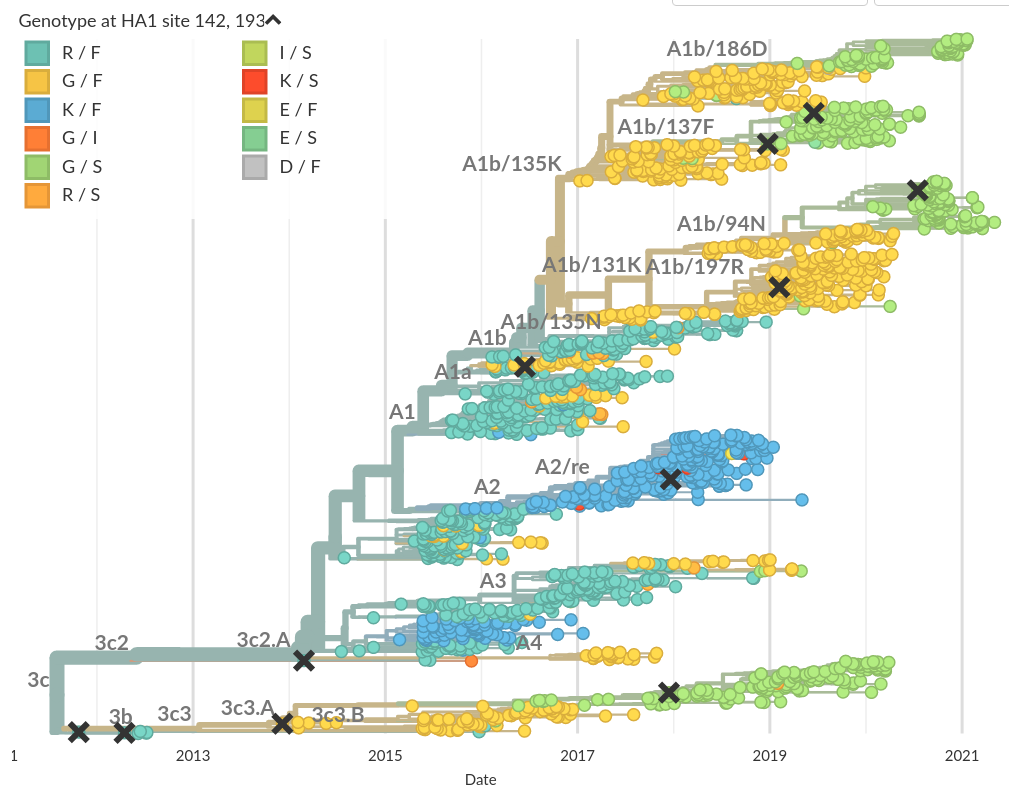
Convergent evolution is common in RNA viruses
Influenza A/H3N2:
4-fold convergence at positions 142 and 193 of HA1
- Convergent evolution is a tell-tale sign of adaptation
(hitting the same site by chance is unlikely) - But: RNA viruses are not mutation limited
- In suitable environments, homoplasies are common
- Variation at homoplasic positions is often selected
→ but success depends on background
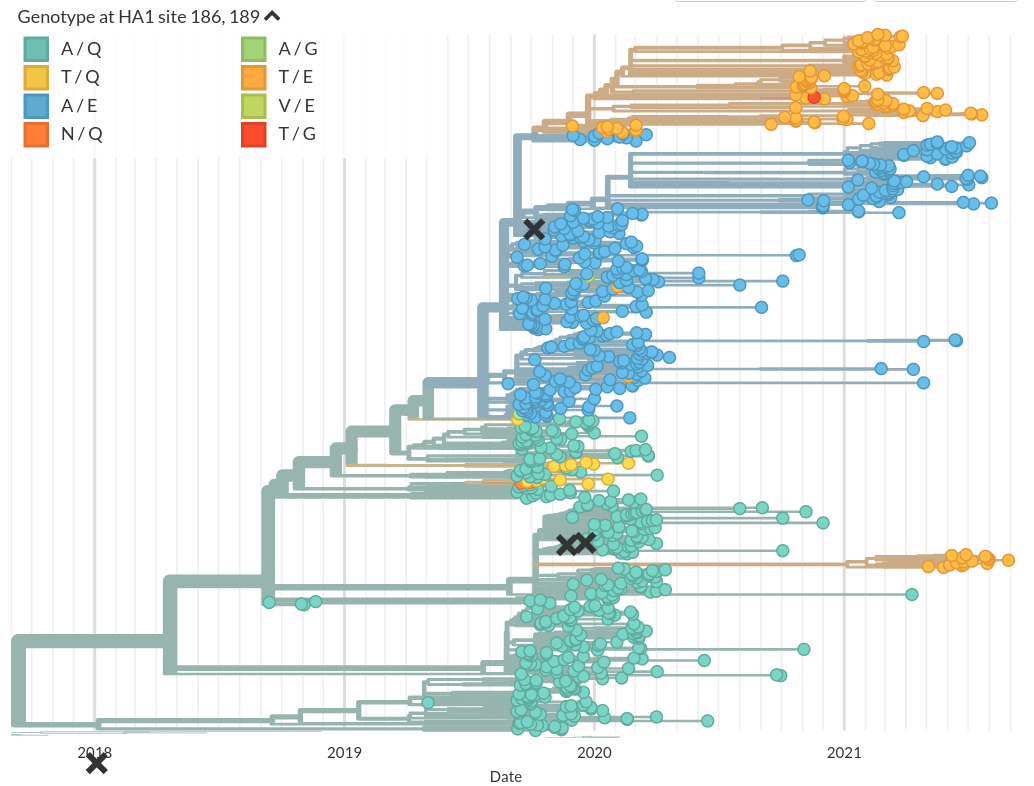
Convergent evolution is common in RNA viruses
A/H1N1pdm:
- Convergence at positions 186 and 189
- Ferret serology is rather insensitive to these changes, but very sensitive to 156K
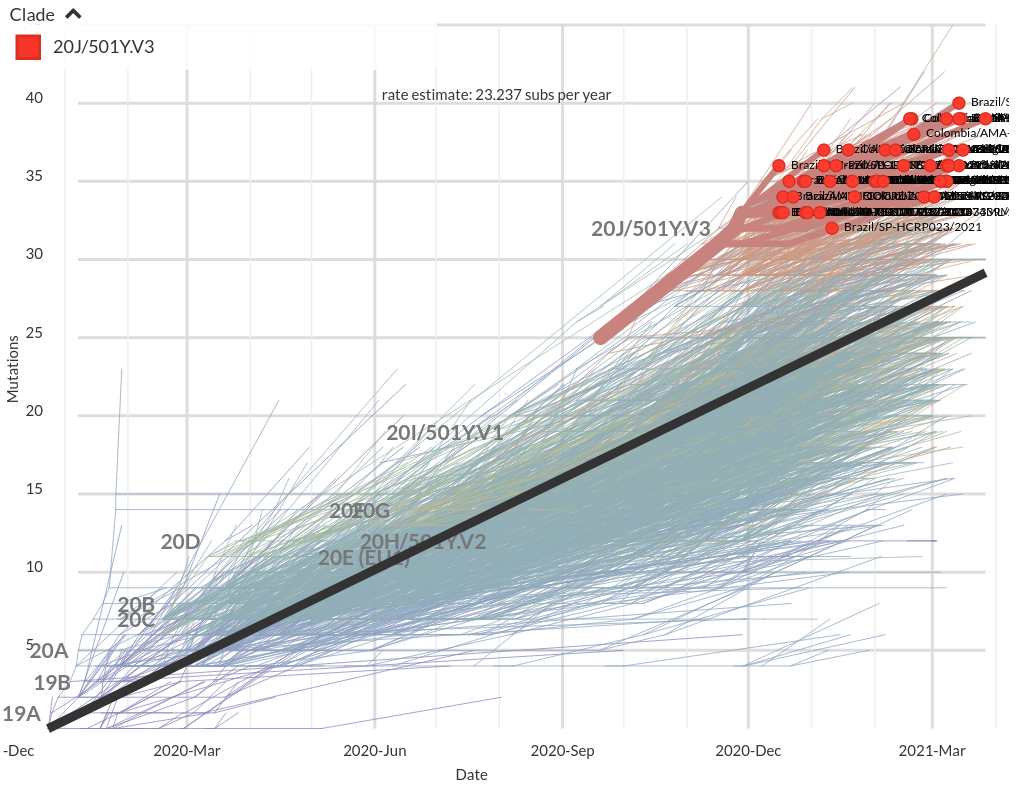
Mutational signatures
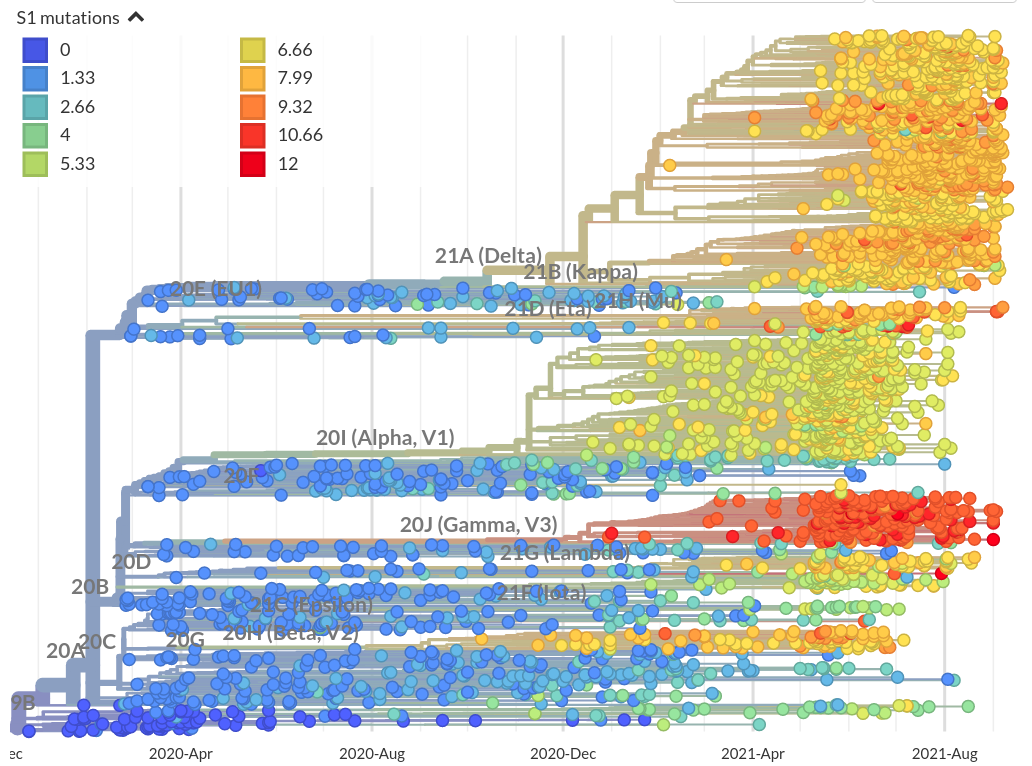
Number of amino acid changes in the S1 domain
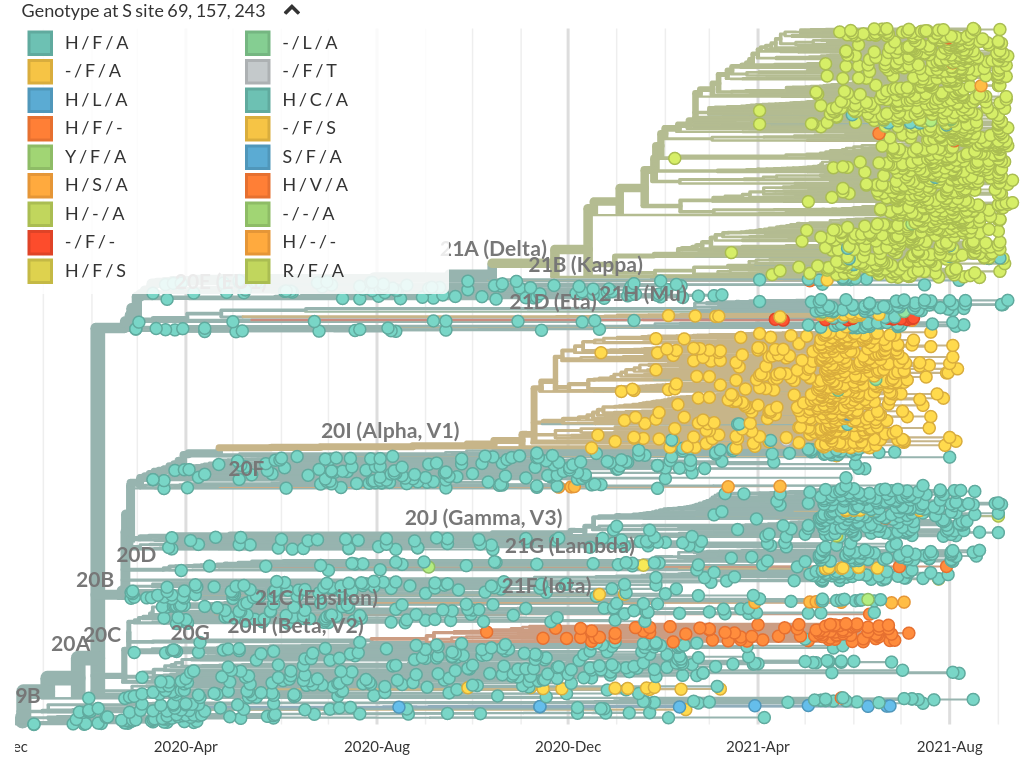
Prominent deletions the spike NTD
Common convergent deletion in NSP6
Growth measures and experimental scores
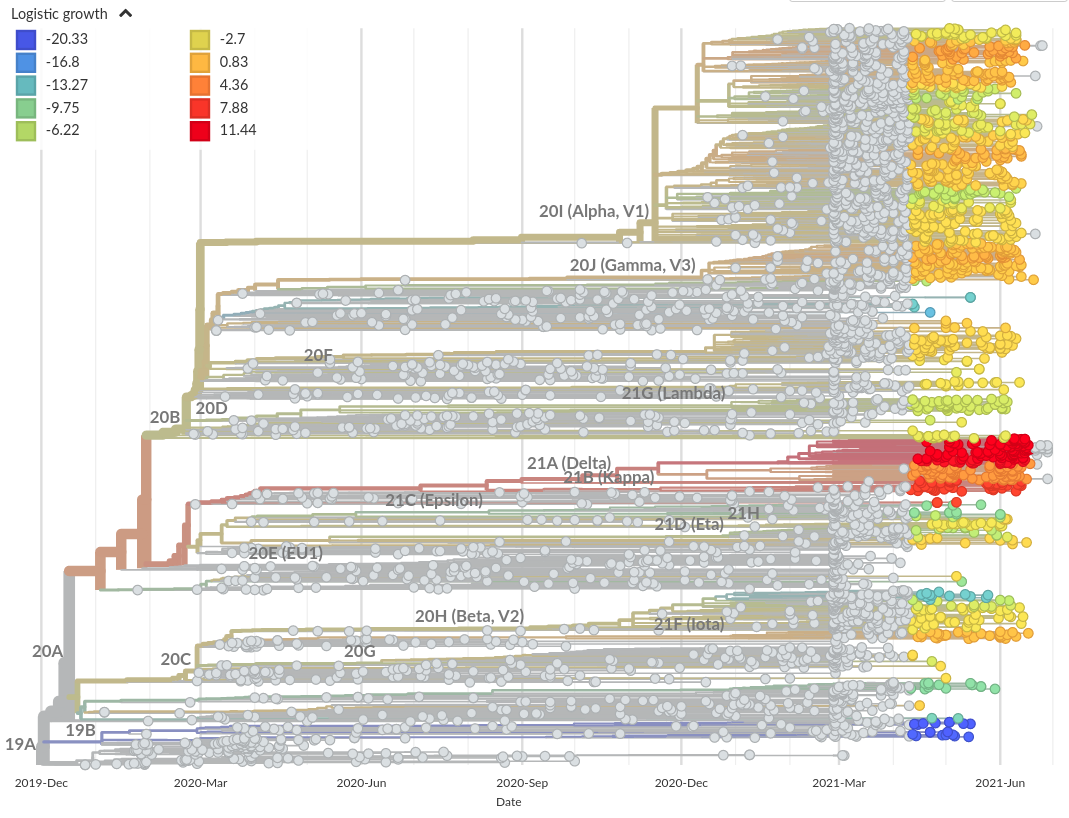
Recent growth rate
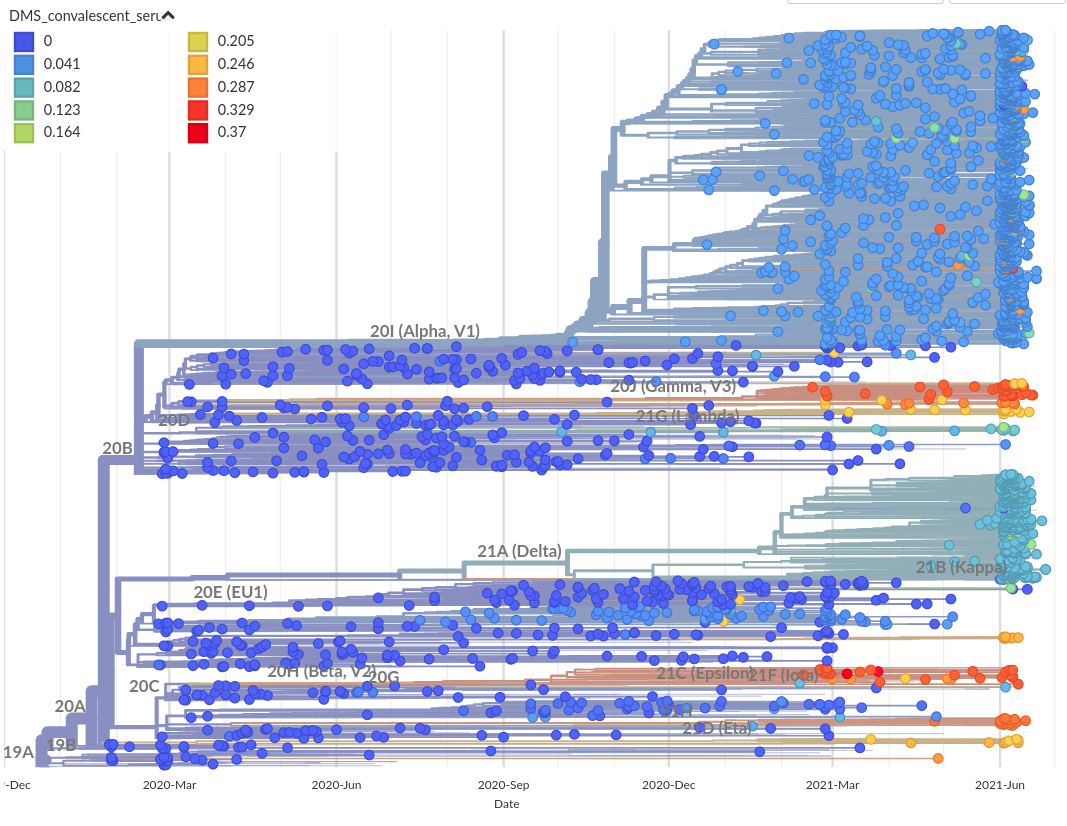
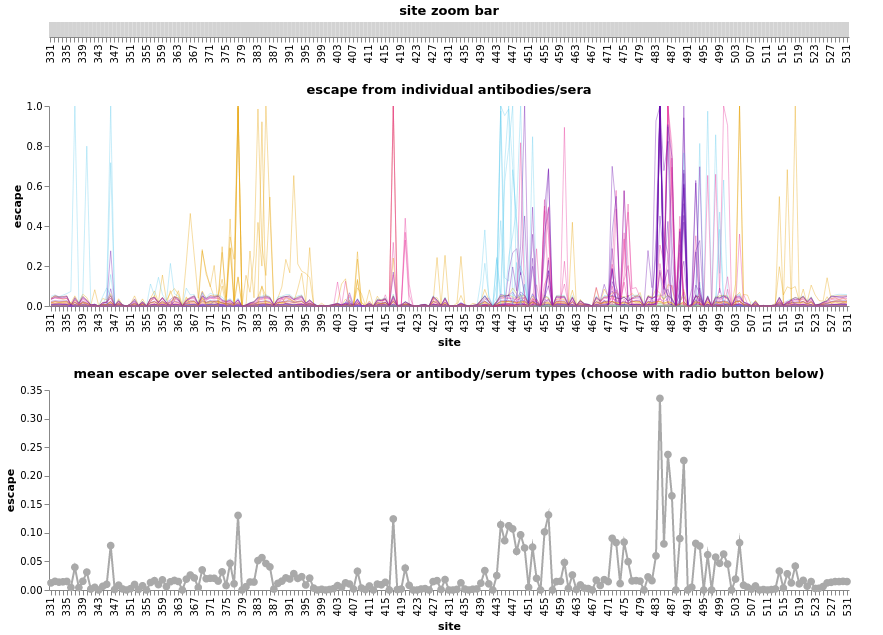
RBD escape based on deep mutational scanning.
Allie Greaney (Jesse Bloom's lab)
Deep mutational scanning for RBD mutations that affect AB binding and ACE2 binding
- By Tyler Starr, Allie Greaney, Jesse Bloom and colleagues
- Systematic evaluation of all RBD mutations for escape and ACE2 binding
- Monoclonals binding to class 1,2,3,4 epitopes
- Convalescent and vaccine sera to map population escape
- Escape maps of indivual sera and antibodies by Bloom lab
- Escape calculator for retained binding by Jesse Bloom
Conclusion
- Since late 2020, dominance of variants with abundant changes in Spike
- So far, successful variants had increased transmissibility, not immune escape
→ Beta, Gamma, and Mu are driven out by Delta - We expect a gradual shift to selection for immune escape
- Prior to Delta, success of variants was regional. Delta is successful across the globe
 Zhou et al, 2021
Zhou et al, 2021
Acknowledgments
Trevor Bedford and his lab -- terrific collaboration since 2014

especially James Hadfield, Emma Hodcroft, Ivan Aksamentov, Moira Zuber, John Huddleston, and Tom Sibley
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID

