Using open data to track and predict infectious disease
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/202109_MPG.html
Open Source & methods
Open Data
Data sharing in public health emergencies
data owner ↔ public interest
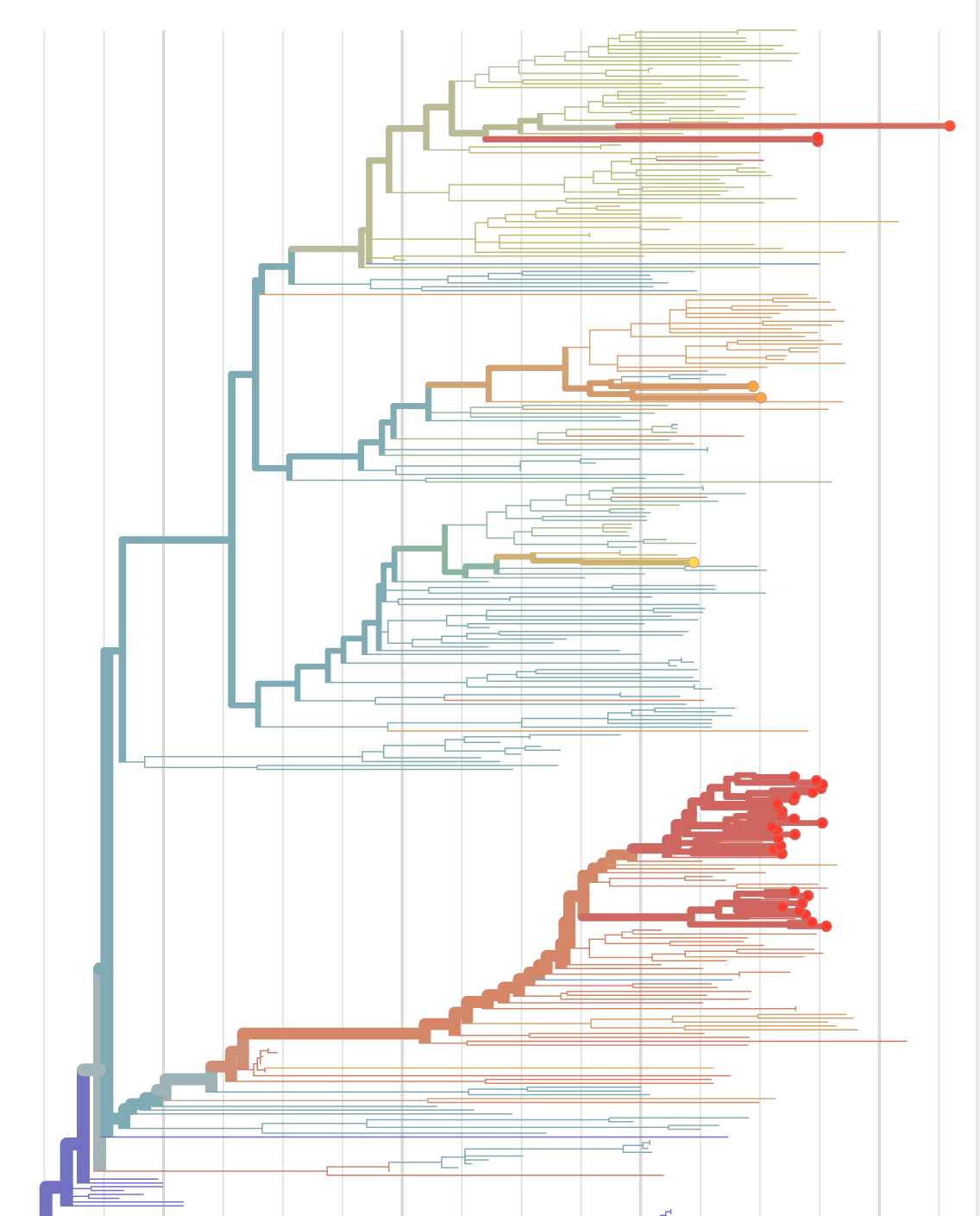
Sequences record the spread of pathogens




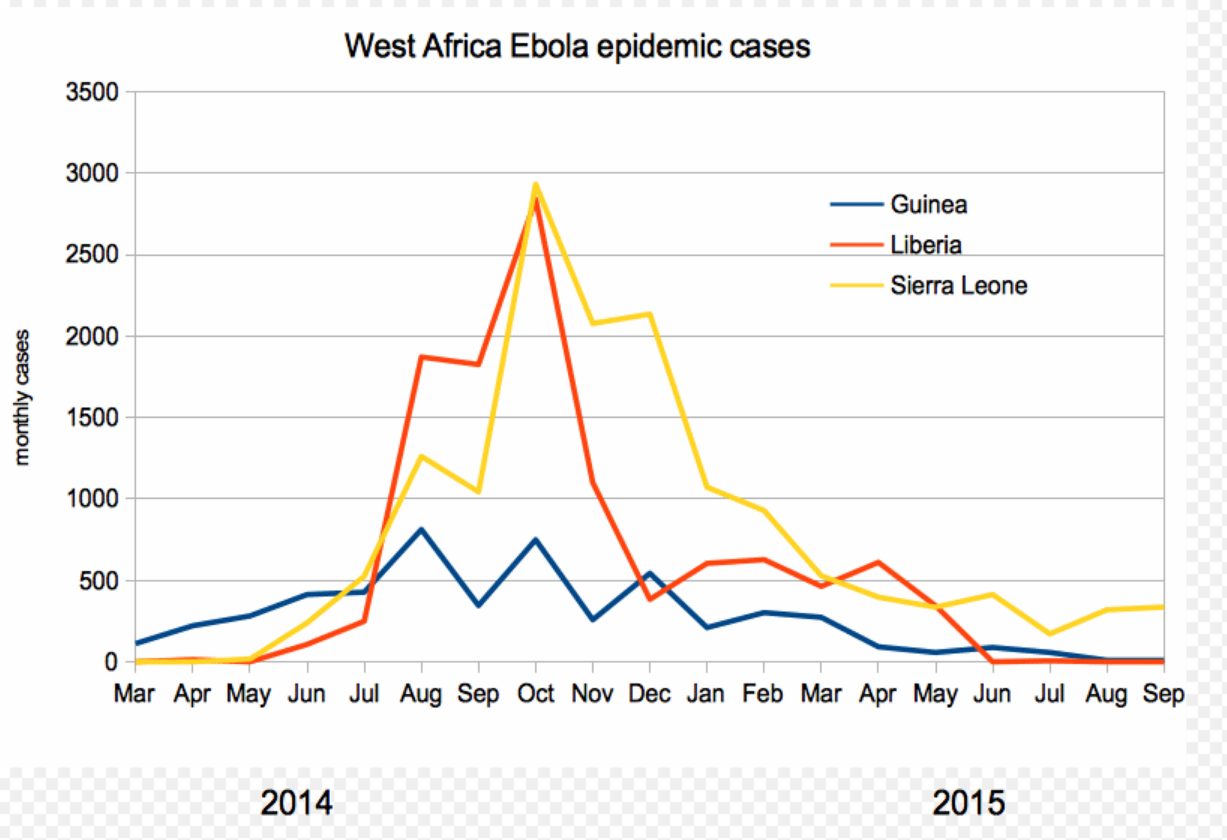
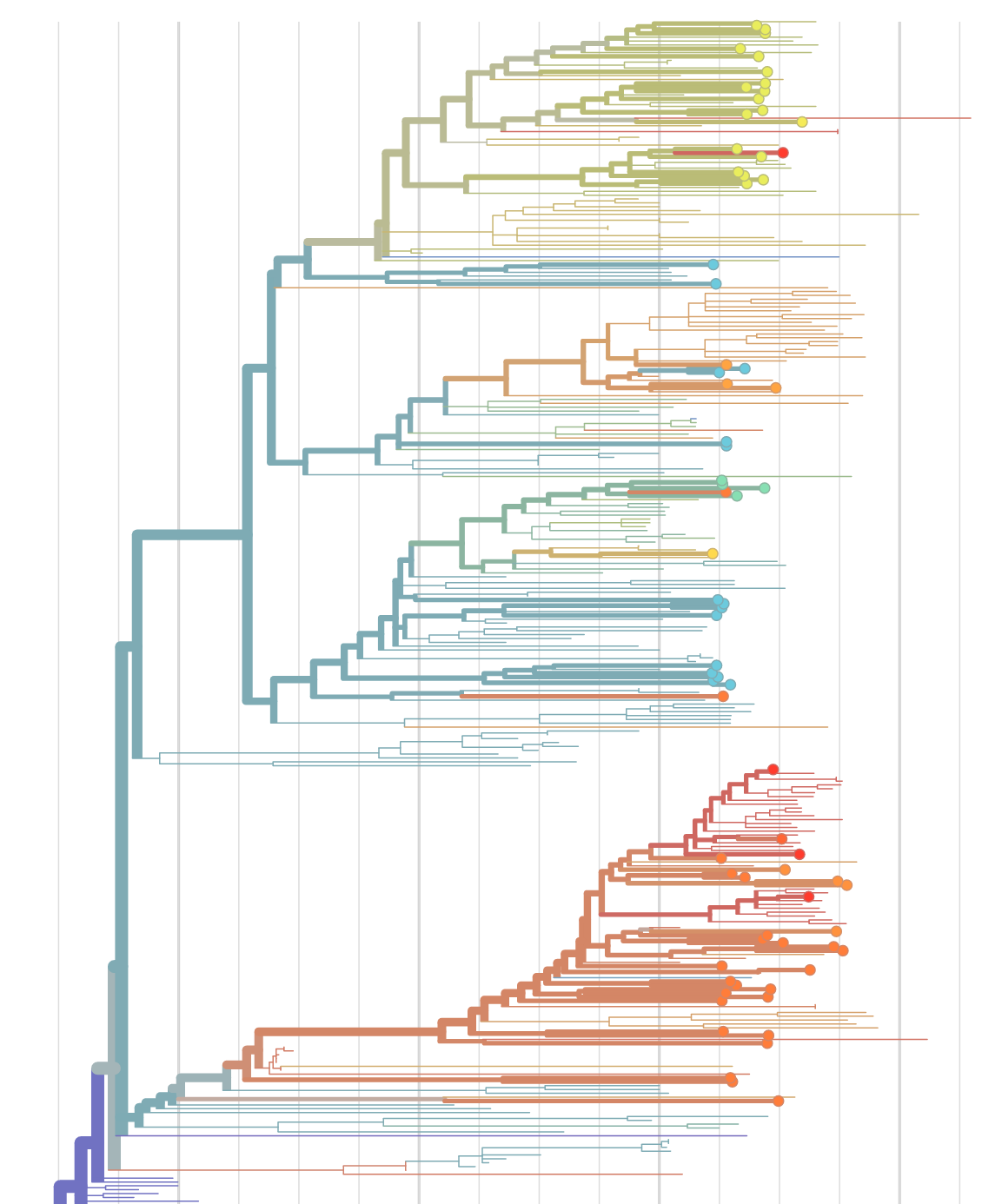
2013-2015 West African Ebola virus outbreak
It took two years to have a detailed understanding of the outbreak. Why did this take so long?
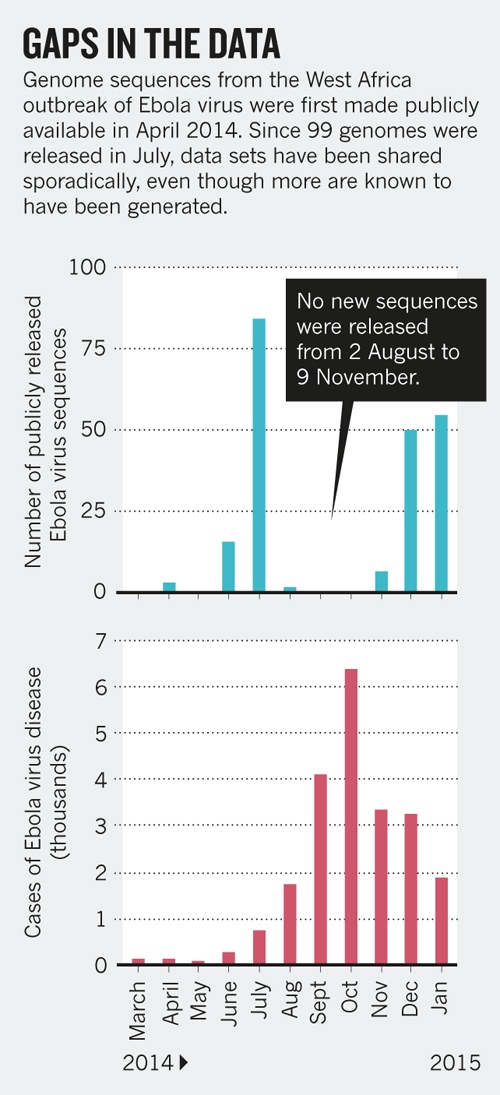
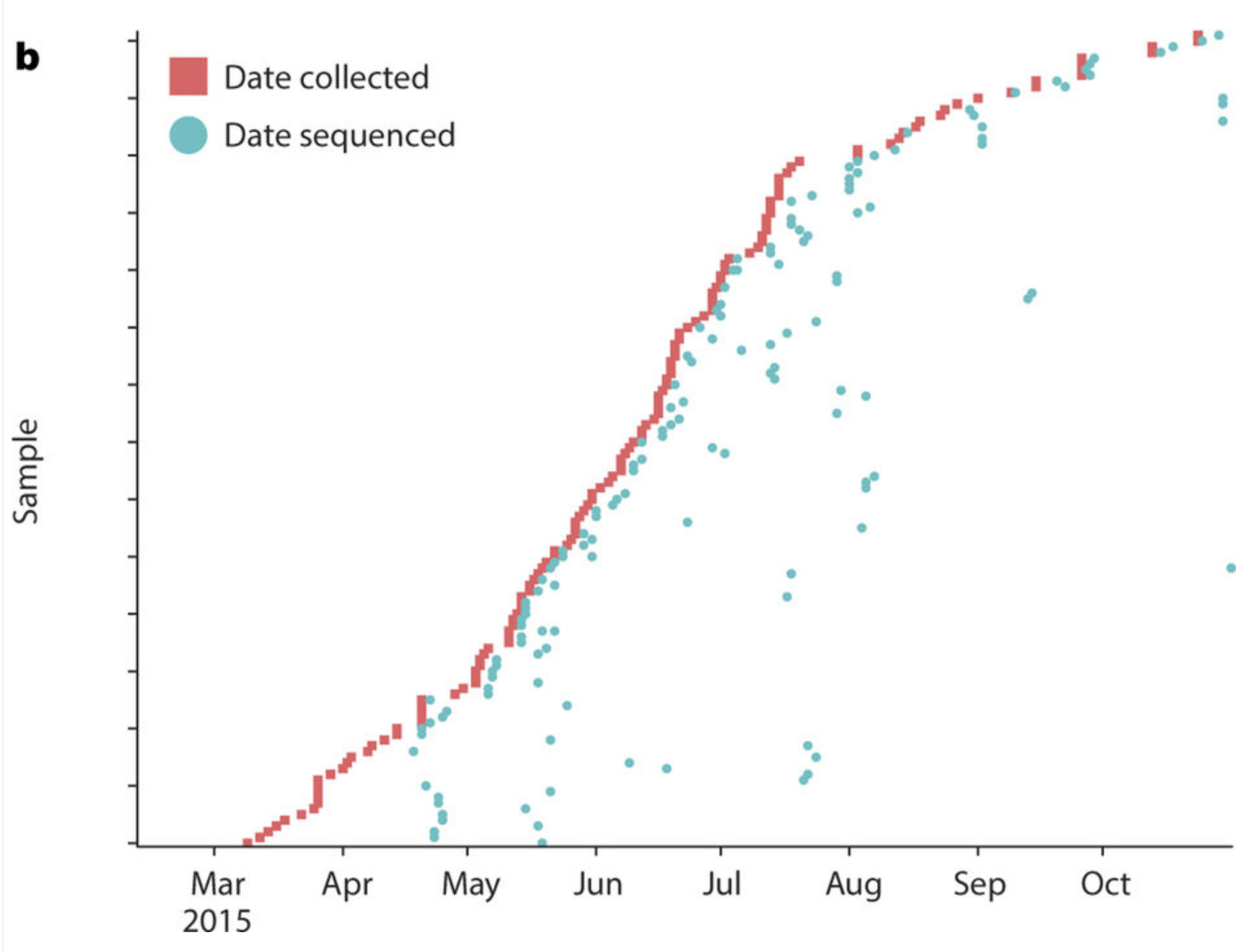
Sharing of Ebola virus sequences
- Baize et al.: Samples collected in March 2014, sequences in GenBank within a month
- Gire et al.: released data as it was generated
- Followed by a long gap...
- Early insights into transmission dynamics are important
- Sharing has to be immediate -- not upon publication
Recurring problem
- 2002 Severe acute respiratory syndrome (SARS)
- 2003 H5N1 influenza outbreak. Some countries stopped sharing any data
- 2013-2015 Ebola virus outbreak in West Africa
- 2014-2016 Zika Virus outbreak: Controversies about attribution and reuse
- 2020- SARS-CoV-2: remarkably rapid data sharing, but continued discussions
Different disease -- different scientists and institutions.
→Lessons need to be relearned.
Barriers to data sharing: scientists
- Privacy of study participants
- Fear of being scooped/ensure maximal return
- Secondary analysis perceived as freeloading: "data parasites"
- Don't want to be second guessed
- Release and curation is laborious
- Sloppy records
Also see Smith et al, F1000 2016
Barriers to data sharing: organizations and governments
- Economic consequences of outbreaks (tourism, agriculture)
- Conflicts between high and low/middle income countries
- Concerns about IP and commercial exploitation
- Legislative/regulatory barriers
See also: van Panhius et al, BMC Public health 2014
Overcoming Barriers
Open vs restricted sharing
- Alternatives to GenBank: GenBank is "public domain", no requirement to credit data producers
- GISAID/EpiFlu: sign-up and agree to terms and conditions

- virological.org: Platform for sharing and discussing molecular epidemiology
- Explicit data reuse terms
- Outline planned projects in white-paper
- Caveat: Very difficult to enforce...
See Bogner et al, 2006
Building Trust
- GISAID coordinates Influenza and SARS-CoV-2 data sharing and curation
- Andrew Rambaut coordinated Ebola virus data sharing
- During the EBV outbreak, WHO and journals explicitly encouraged data sharing
See also Smith et al, F1000 2016
Make sharing easy and provide incentives!
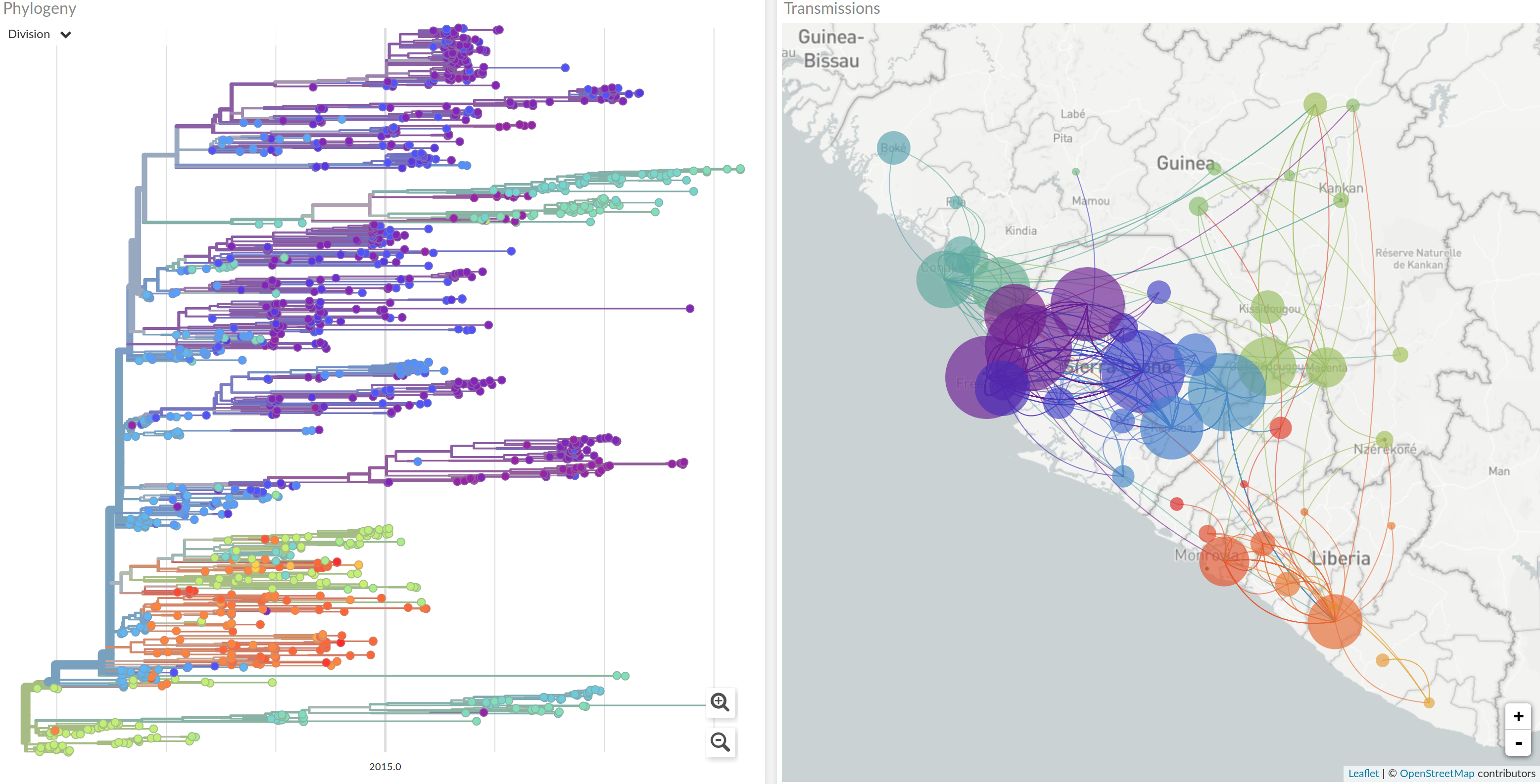
Nextstrain.org
joint work with Trevor Bedford & his lab
code at github.com/nextstrain
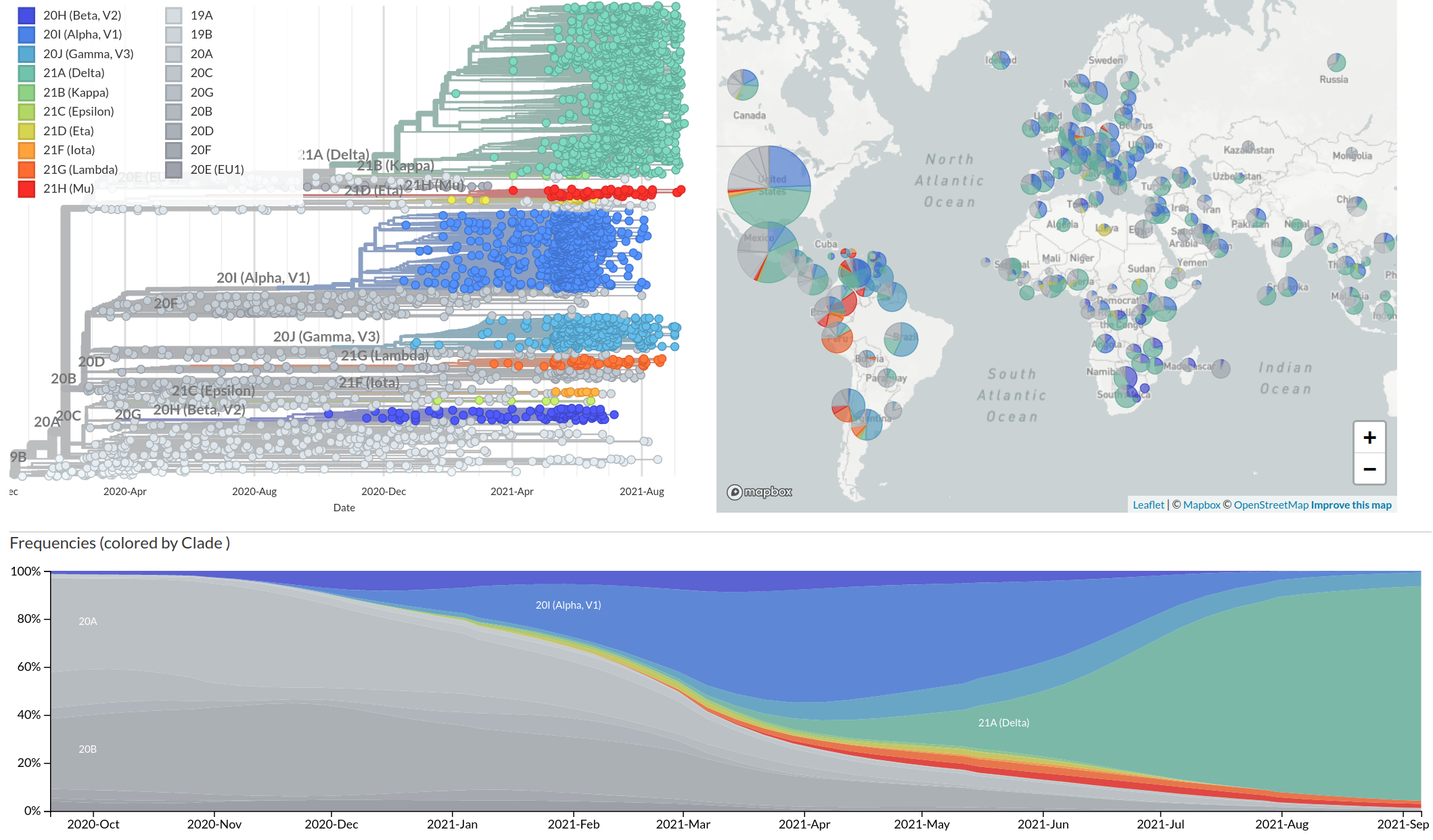
nextstrain.org
- Global analysis provides context for new sequences
- Once nextstrain became the largest collection of Ebola/Zika sequences, everybody wanted their sequences on nextstrain
- We take care to highlight the scientists contributing data
- Link to original source, rather than reshare
- nextstrain.org/zika?f_authors=Metsky et al
Open source: github.com/nextstrain
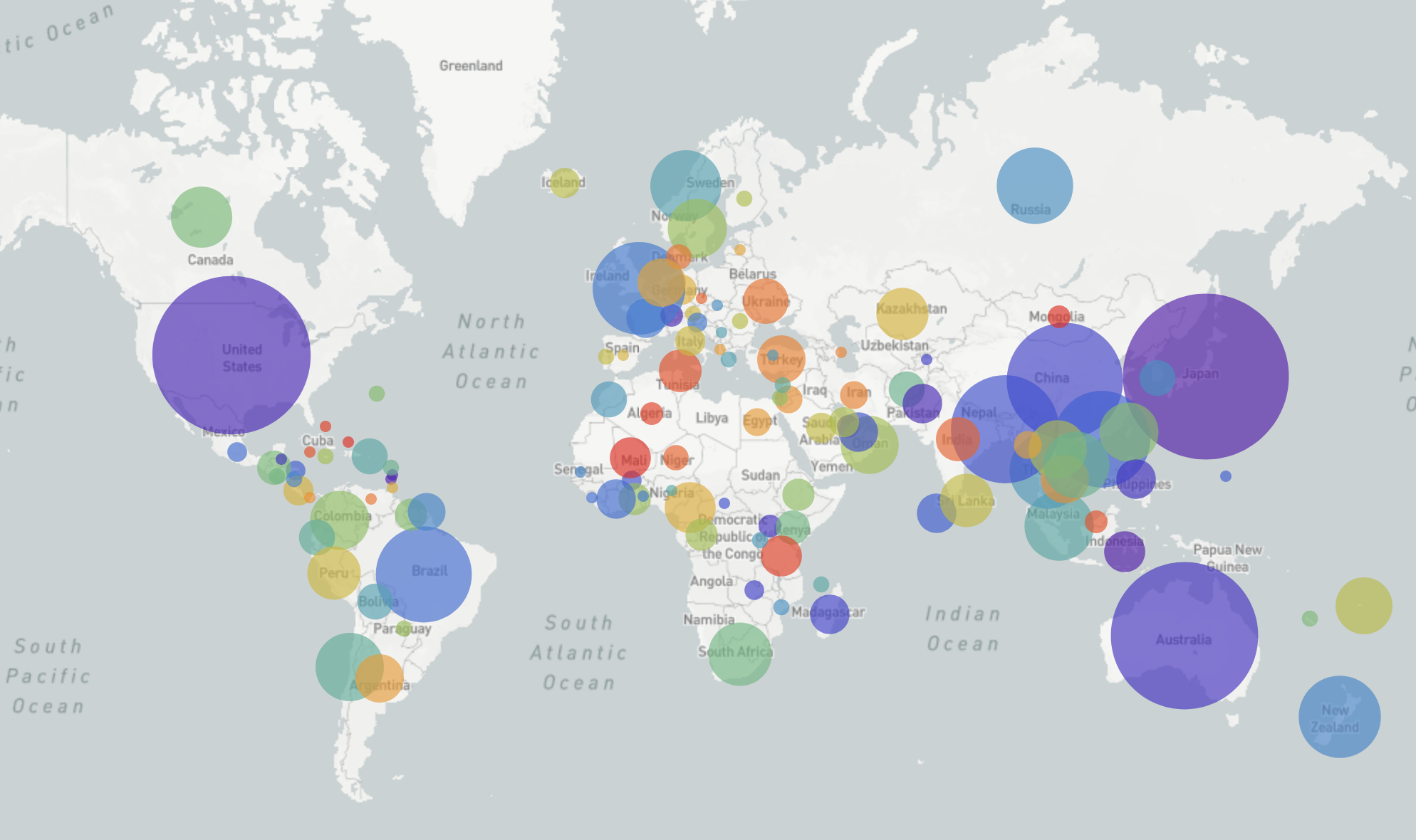
Open Data makes wonderful things possible
- Most coronavirus trackers are only possible because somebody decided to share data in a machine readable form.
- Publicly generated data (maps, air quality, weather, disease circulation, demographics, etc) should be open whenever legally possible.
- OurWorldinData.org is a fantastic example
Summary and outlook
- In public health emergencies, immediate data sharing is imperative
- Response has to be fast
→ pre-existing trusted framework to share data
→ rapid and readily deployable analysis tools - We need incentives for high quality data sets
- We need better ways to credit data producers
→ data citations - We should rethink licensing models of data bases.
- Wider acceptance of preprints should help
→ establishes priority and a citation hook
Acknowledgments
Trevor Bedford and his lab -- terrific collaboration since 2014

especially James Hadfield, Emma Hodcroft, Ivan Aksamentov, Moira Zuber, John Huddleston, and Tom Sibley
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID