Real-time tracking of SARS-CoV-2 using Nextstrain
Richard Neher
Biozentrum & SIB, University of Basel
slides at neherlab.org/202109_PAHO.html
Available data on Jan 26
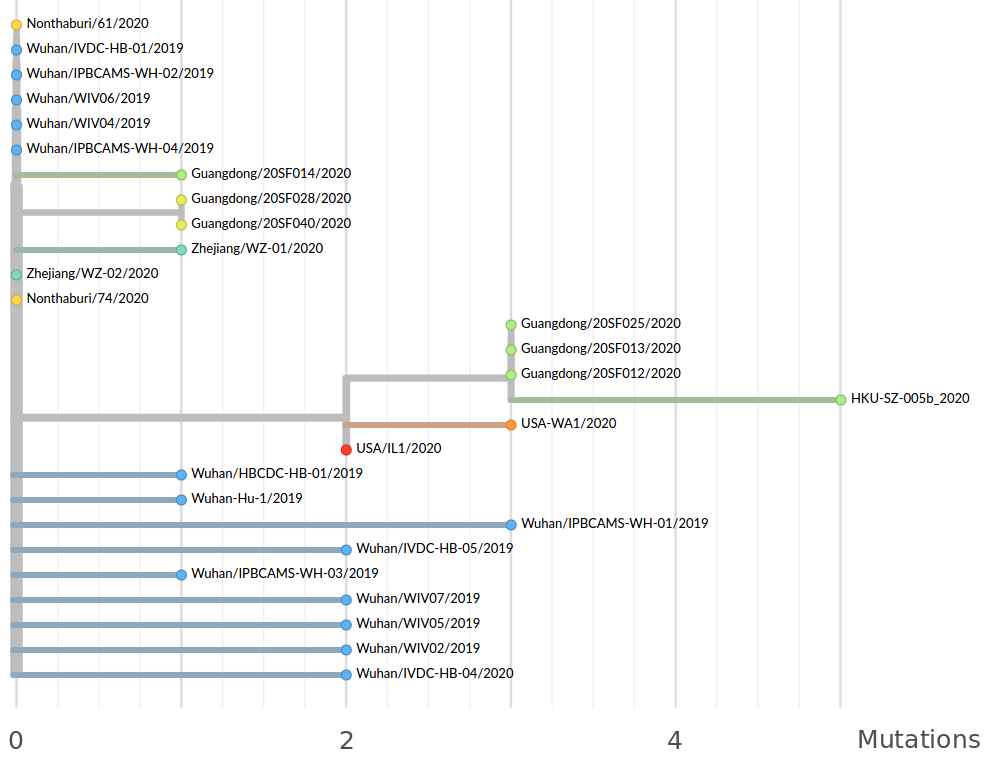
Early genomes differed by only a few mutations, suggesting very recent emergence
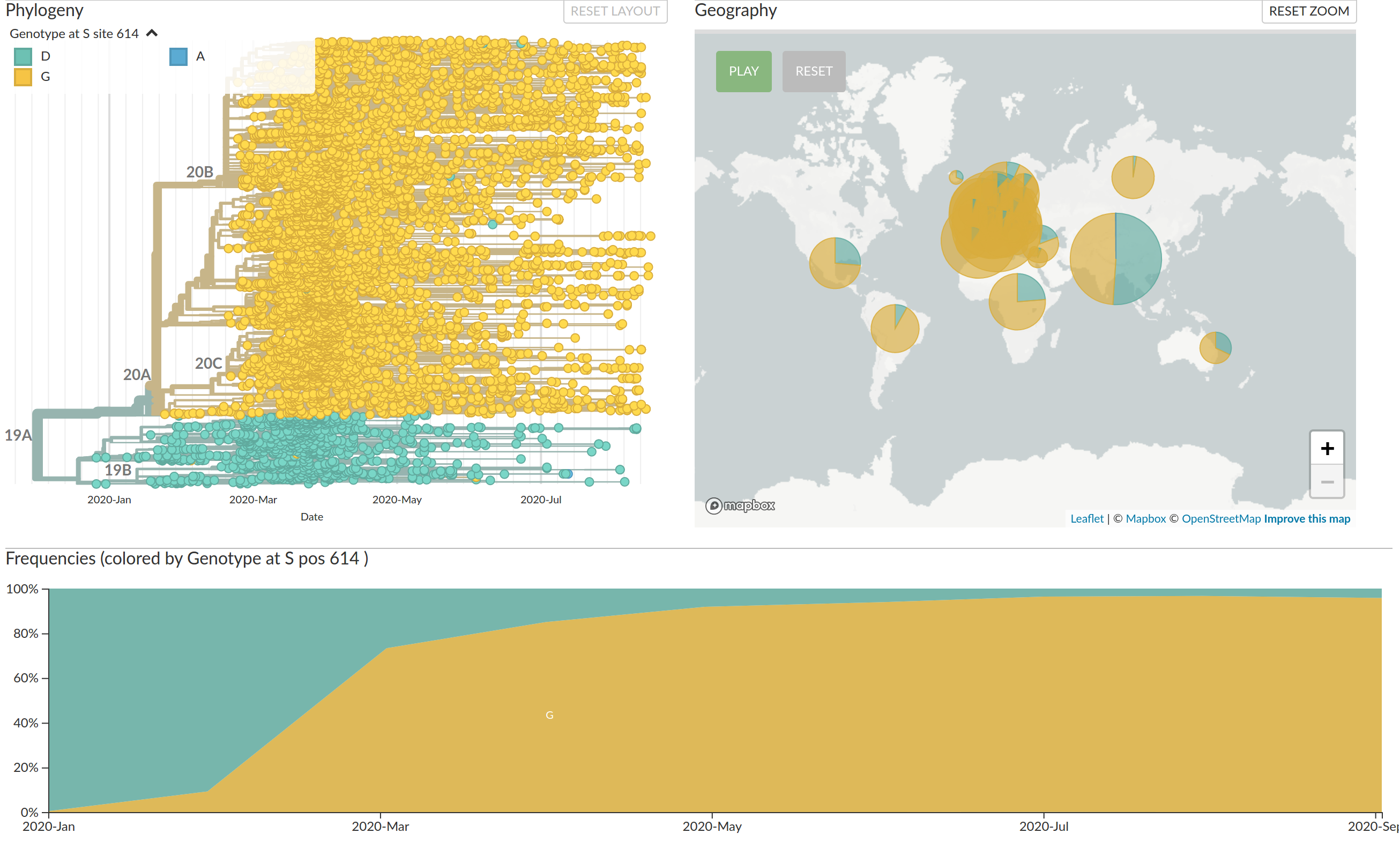
>3.5 million sequences
Interactive part on Nextstrain
- Nextstrain started in 2014 -- before the pandemic, too much data wasn't a problem
- A whole eco-system of tools and trackers has emerged since
- cov-spectrum
- Usher
- outbreak.info
- cov2tree.org
Tracking variants turned out to be crucial
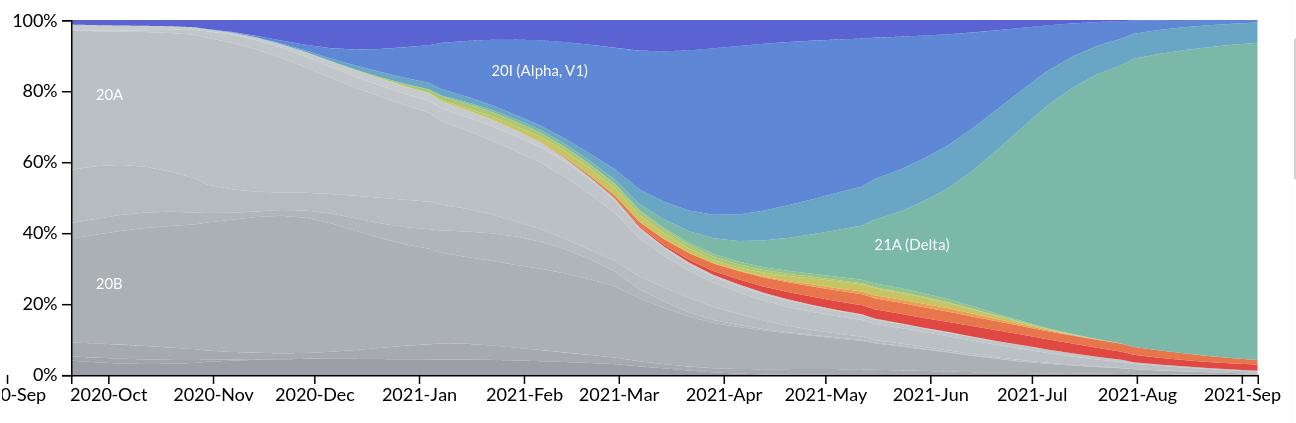
Emergence and dominance of VoCs
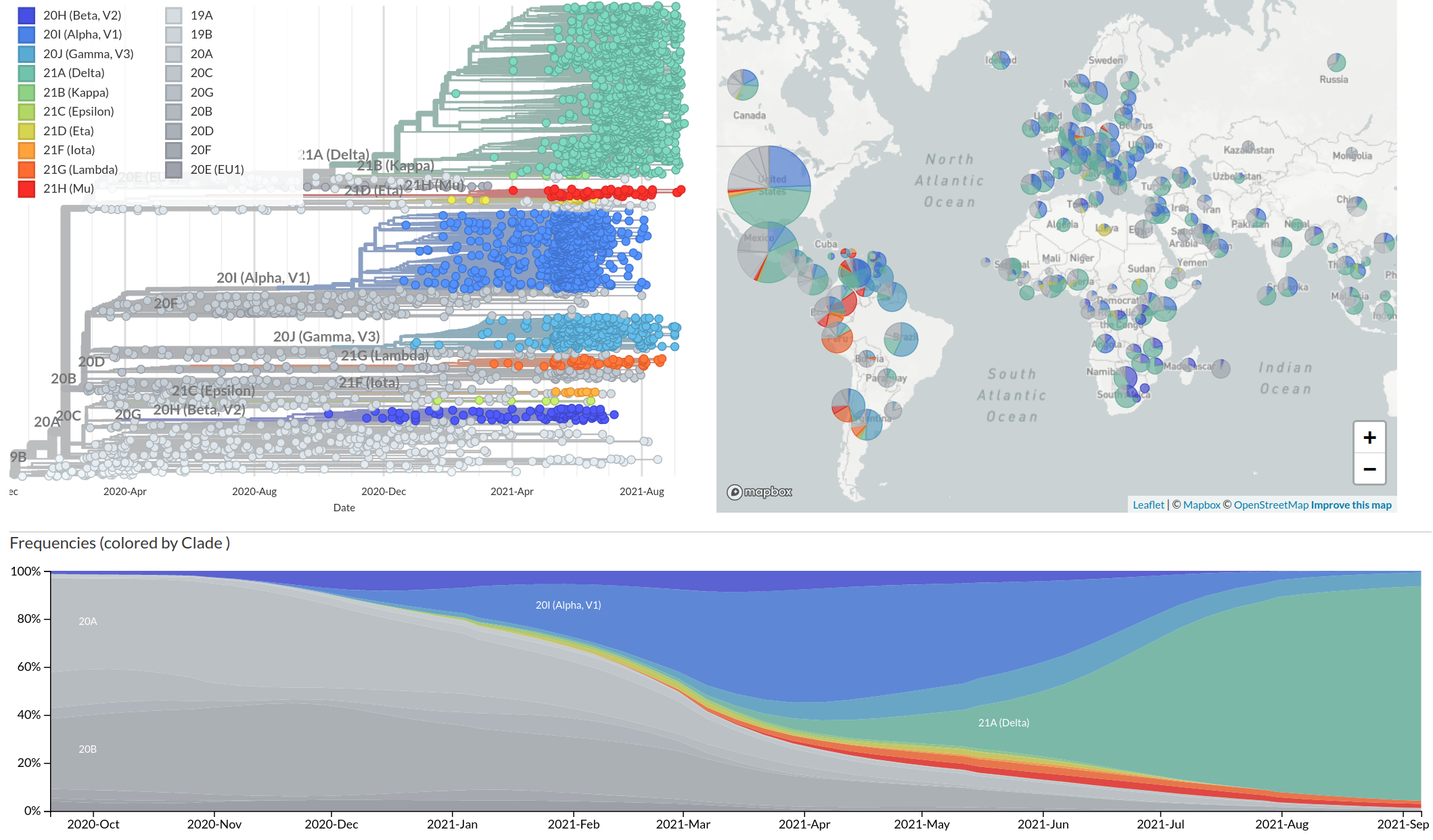
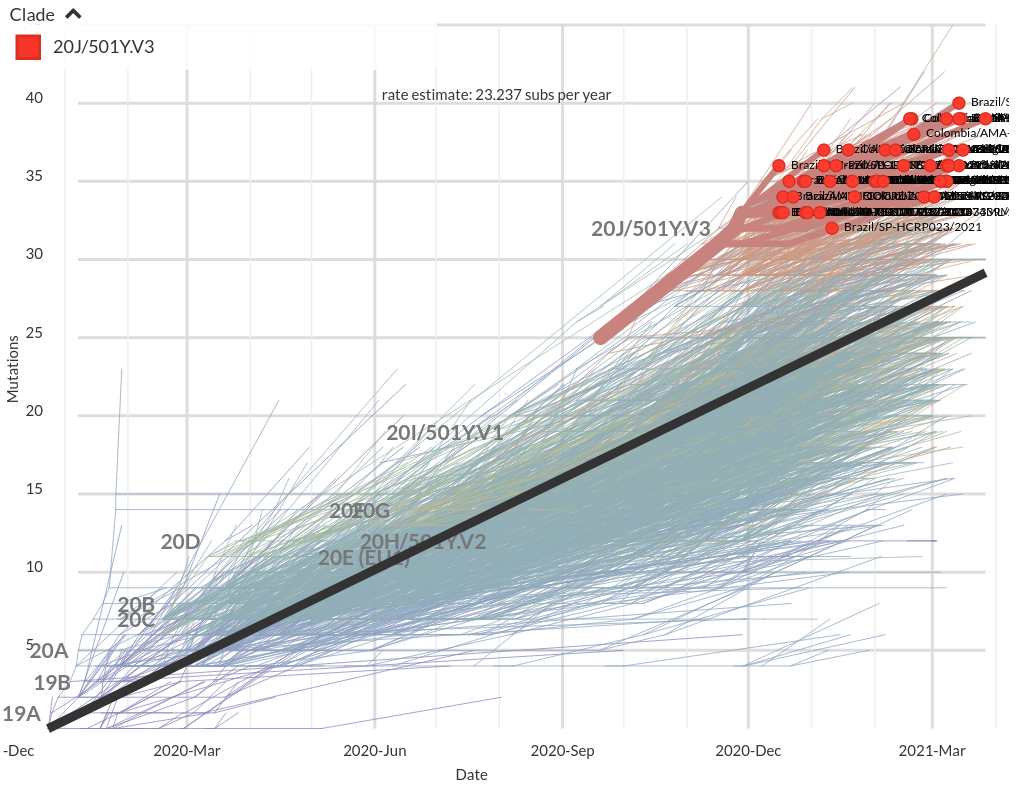
VoCs have more mutations than expected...
 nextstrain
nextstrain
VoCs and VoIs: rapid converging evolution
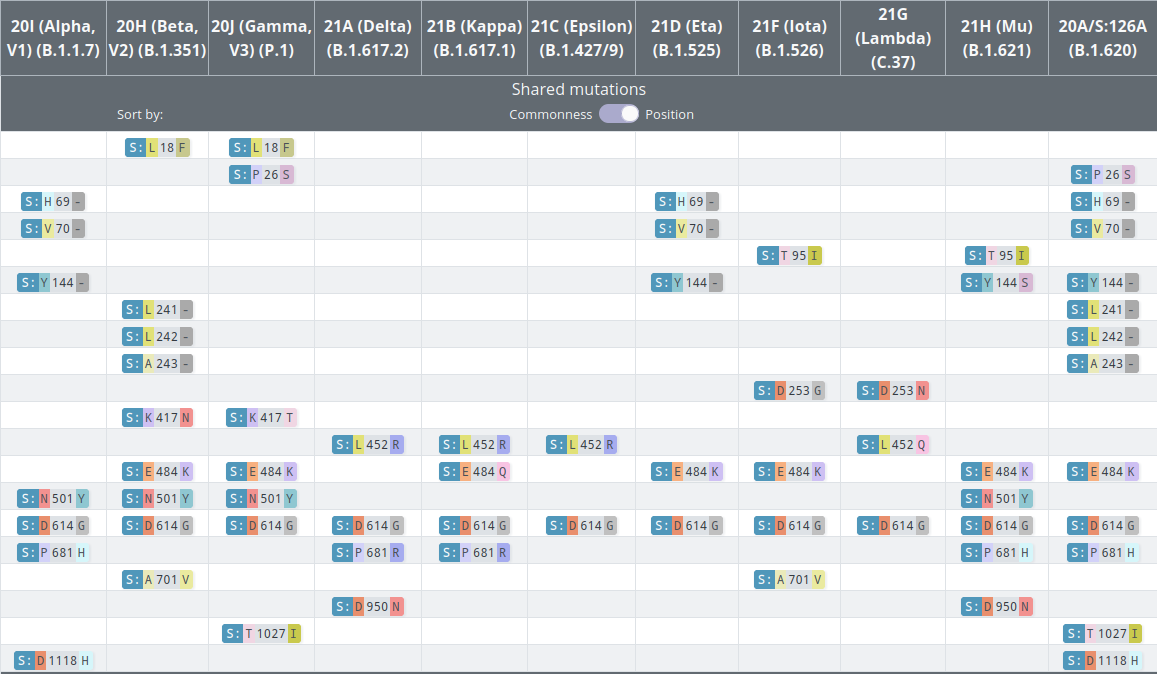 covariants.org by Emma Hodcroft
covariants.org by Emma Hodcroft
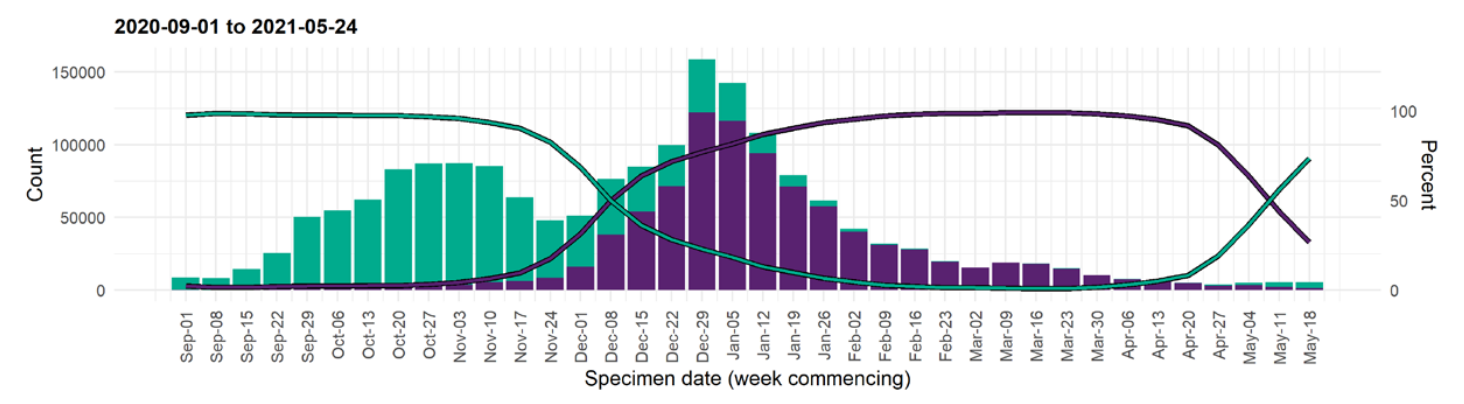
VoCs have been replacing other strains quickly
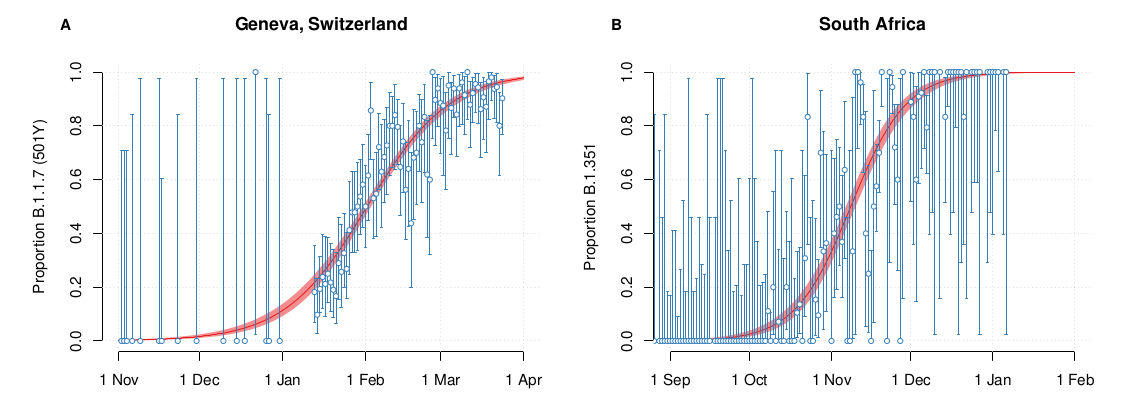
 S-gene positivity in the UK
S-gene positivity in the UK
Global
South America
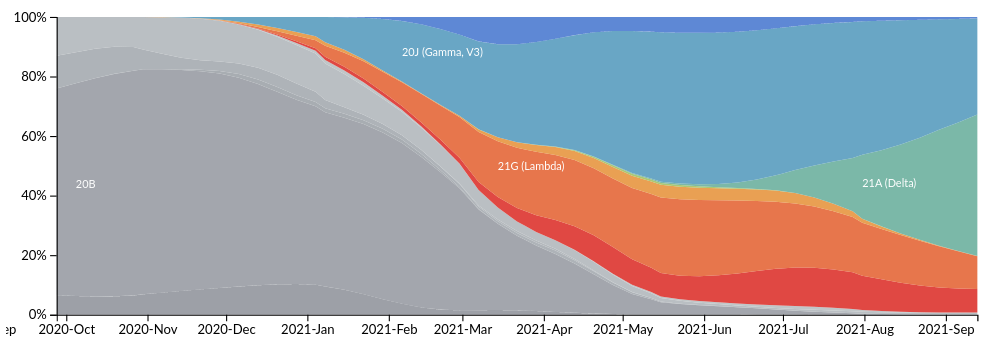
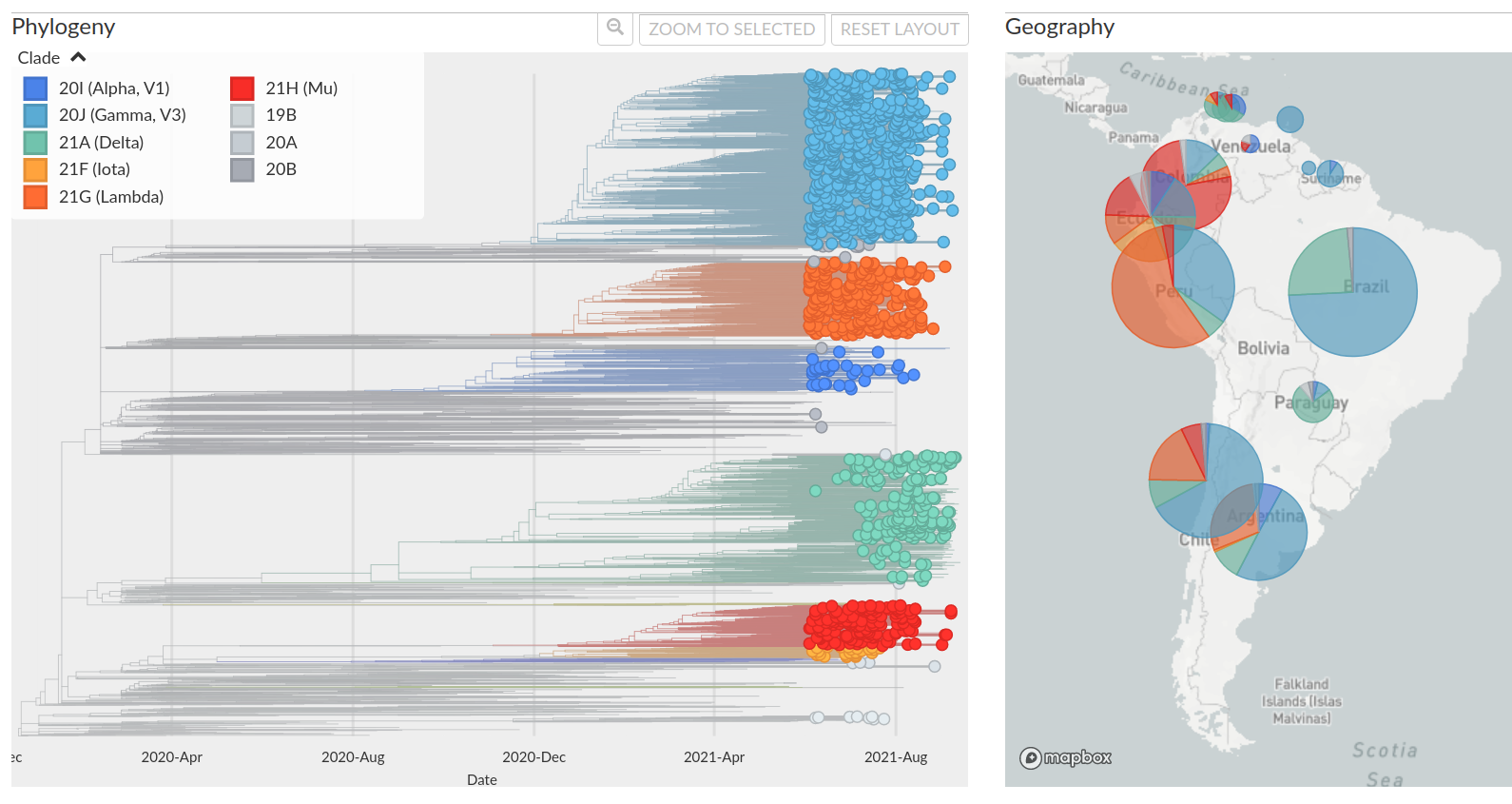
How do we tell early on whether a variant is concerning?
How unusual are the patterns of convergent evolution?
SARS-CoV-2 variants can become dominant without advantage
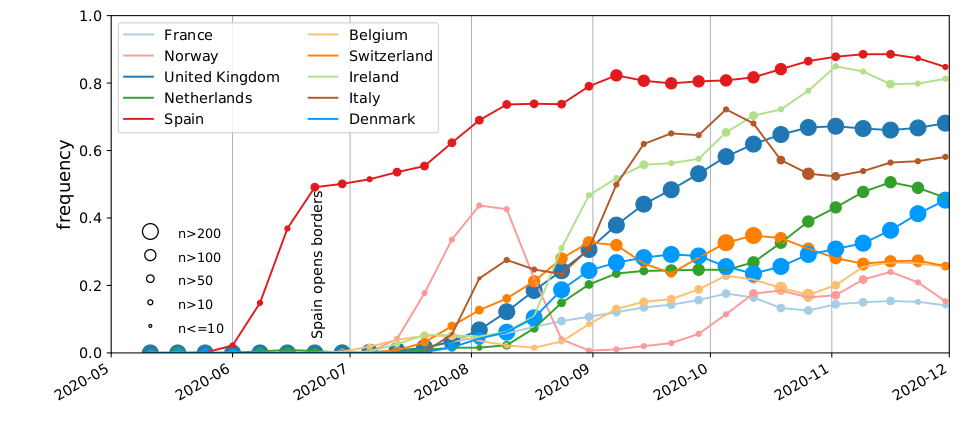
Spanish EU1 diversity was mirrored across Europe
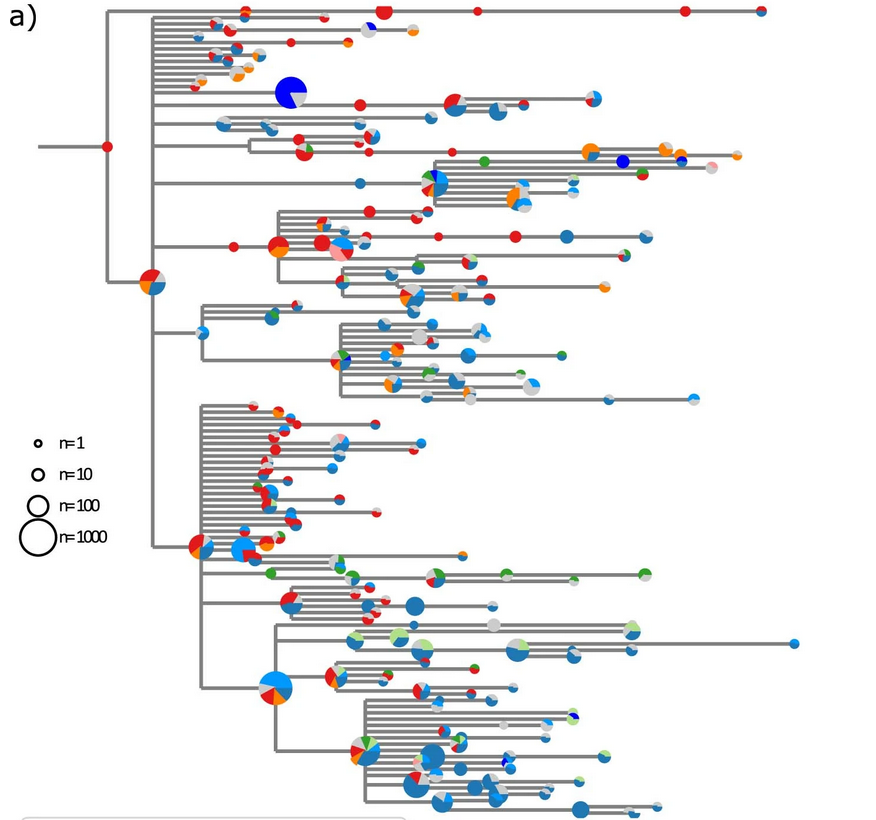

High case numbers in Spain and high travel volume spread the variant
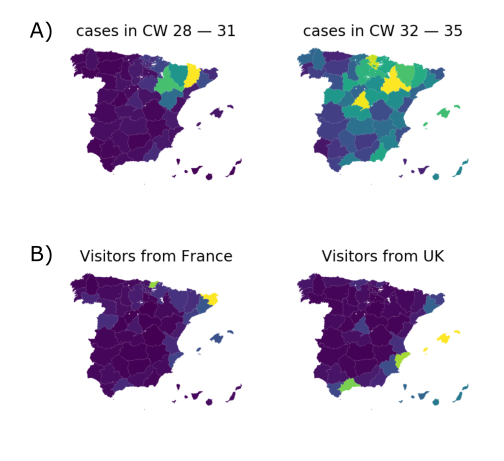
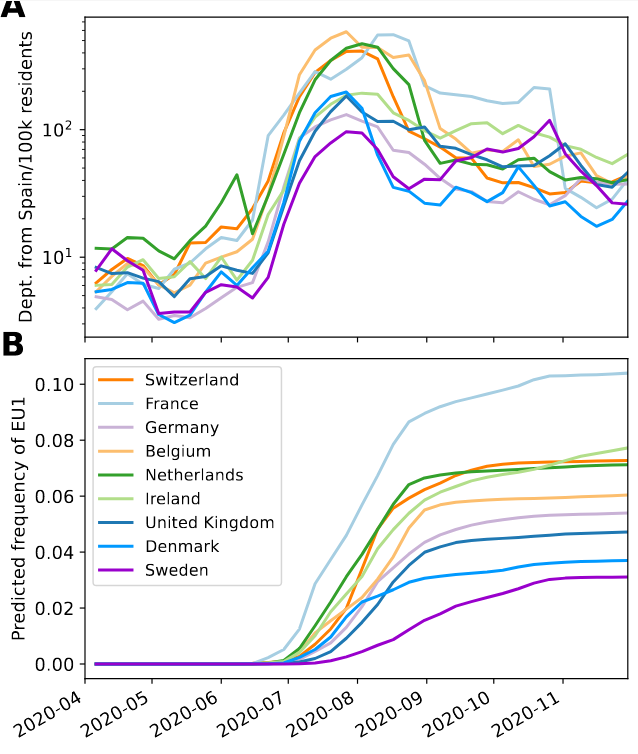
Travel driven variant dispersal and displacement
- High incidence differential and high travel volume can shift variant distribution
- Travel associated activities and behavior further increases impact
- Onward spread in traveling demographics can be higher
Mutational signatures
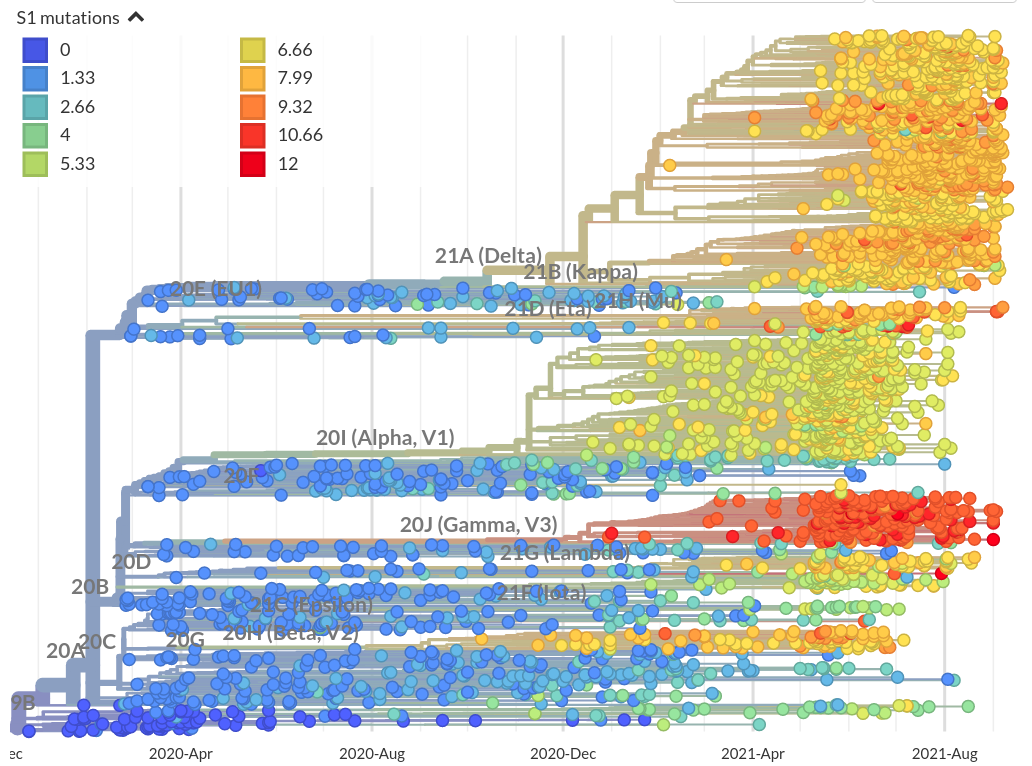
Number of amino acid changes in the S1 domain
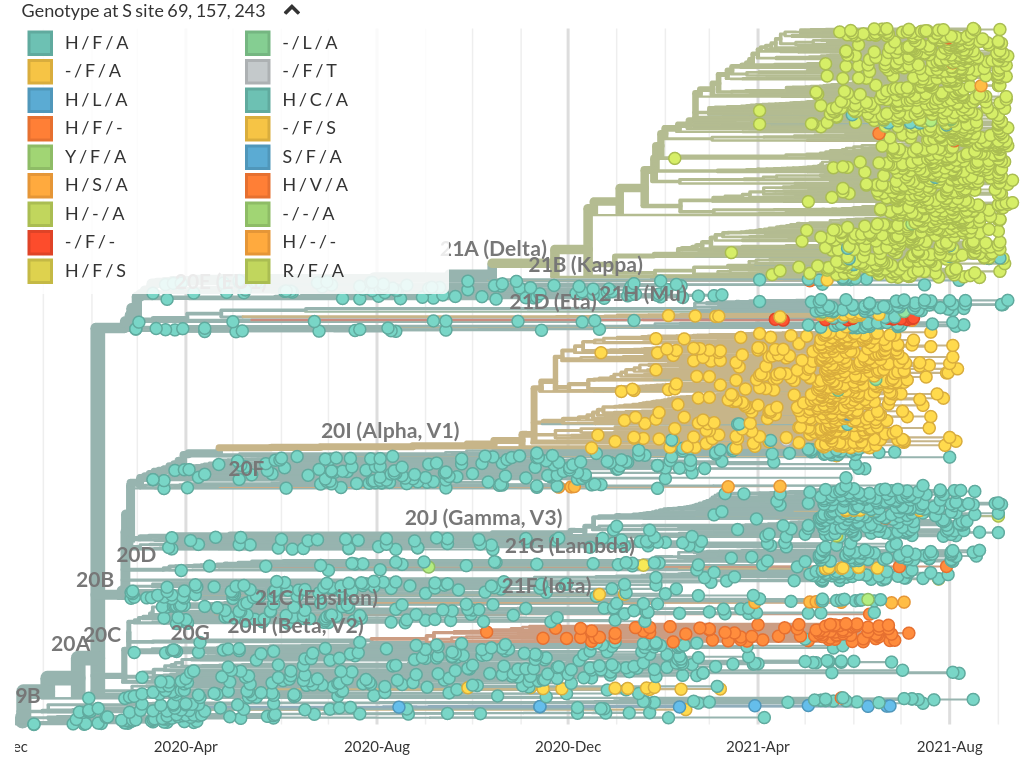
Prominent deletions the spike NTD
Common convergent deletion in NSP6
Growth measures and fitness scores
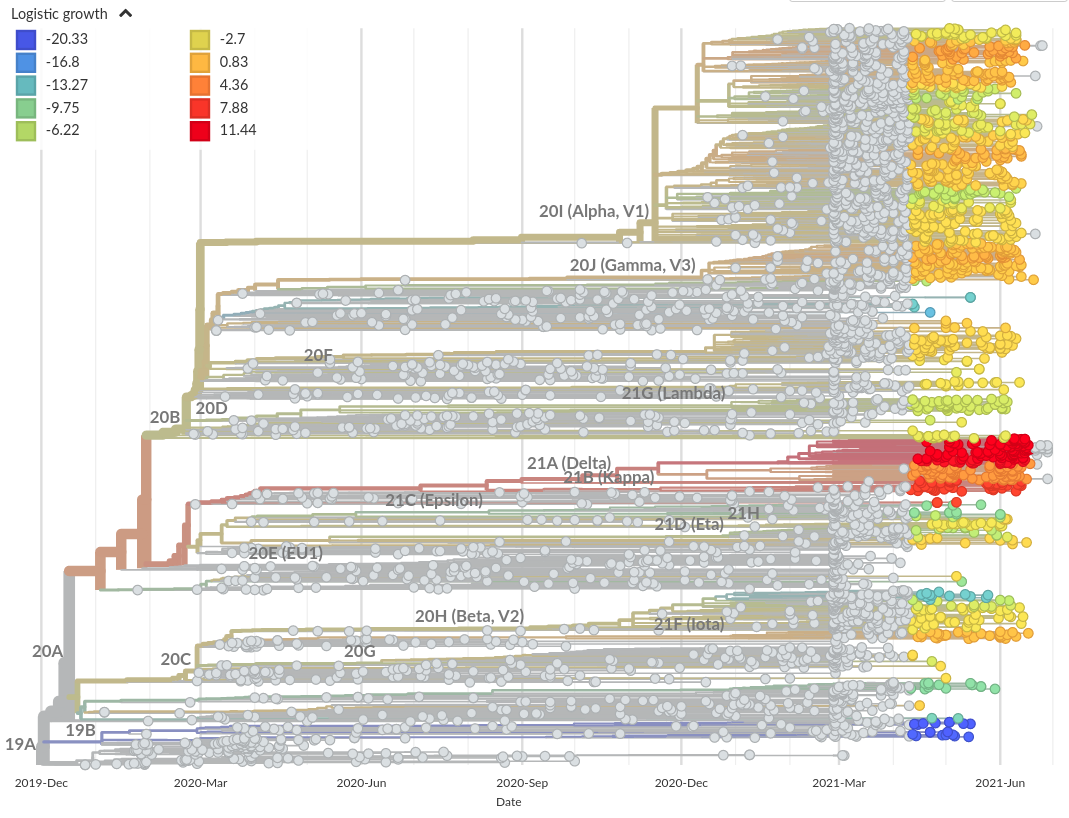
Recent growth rate
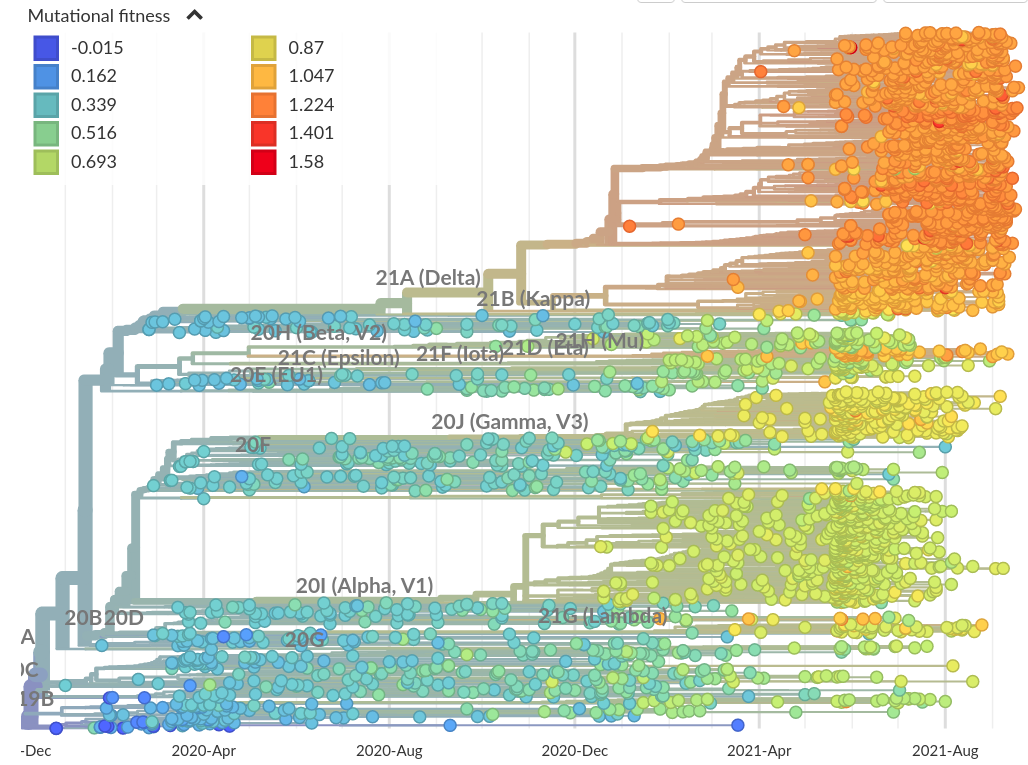
Fitness based on mutational effects by Obermeyer et al
Nextclade
- Within browser alignment and phylogenetic placement
- Calls mutations relative to the reference
- Assigns clades
- Calculates QC metrics
SARS-CoV-2 acknowledgements

.jpg)
- Emma Hodcroft (now in Bern)
- Moira Zuber (Basel)
- Iñaki Comas and Fernando Gonzalez-Candelas, Valencia
- Martina Reichmuth and Christian Althaus (Bern)
- Tanja Stadler, Sarah Nadeau, Tim Vaughan at ETH
- Alberto Hernando and David Matteo at Kido Dynamics
- Jesse Bloom, Katherine Crawford at Fred Hutch
- David Veesler, Alex Walls, Davide Corti, John Bowen at UW

Acknowledgments
Trevor Bedford and his lab -- terrific collaboration since 2014

especially James Hadfield, Emma Hodcroft, Ivan Aksamentov, Moira Zuber, John Huddleston, and Tom Sibley
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID

