Evolution and spread of Influenza, HIV, and SARS-CoV-2
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/202112_CBB.html
Human seasonal influenza viruses

Positive tests for influenza in the USA by week
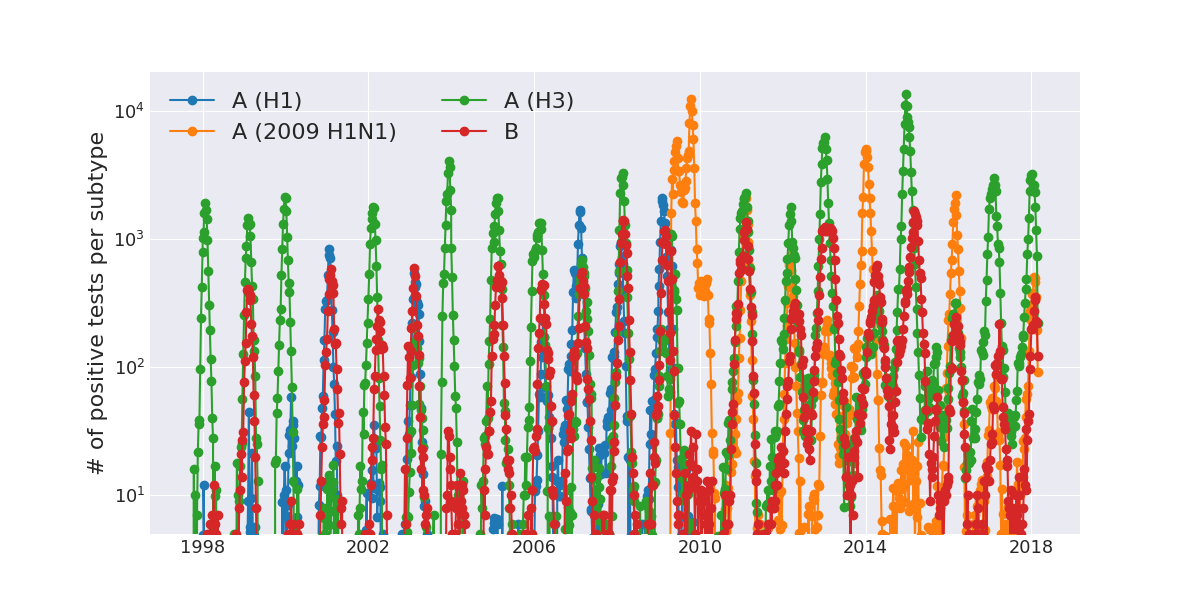 Data by the US CDC
Data by the US CDC
Virus genomes change rapidly through time
A/Brisbane/100/2014
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
A/Brisbane/1000/2015
GGATAATTCTATTAACCATGAAGACTATTATTGCTTT...
A/Brisbane/1/2017
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
... hundreds of thousands of sequences...
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
A/Brisbane/1000/2015
GGATAATTCTATTAACCATGAAGACTATTATTGCTTT...
A/Brisbane/1/2017
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
... hundreds of thousands of sequences...
Genome sequences record the spread of pathogens




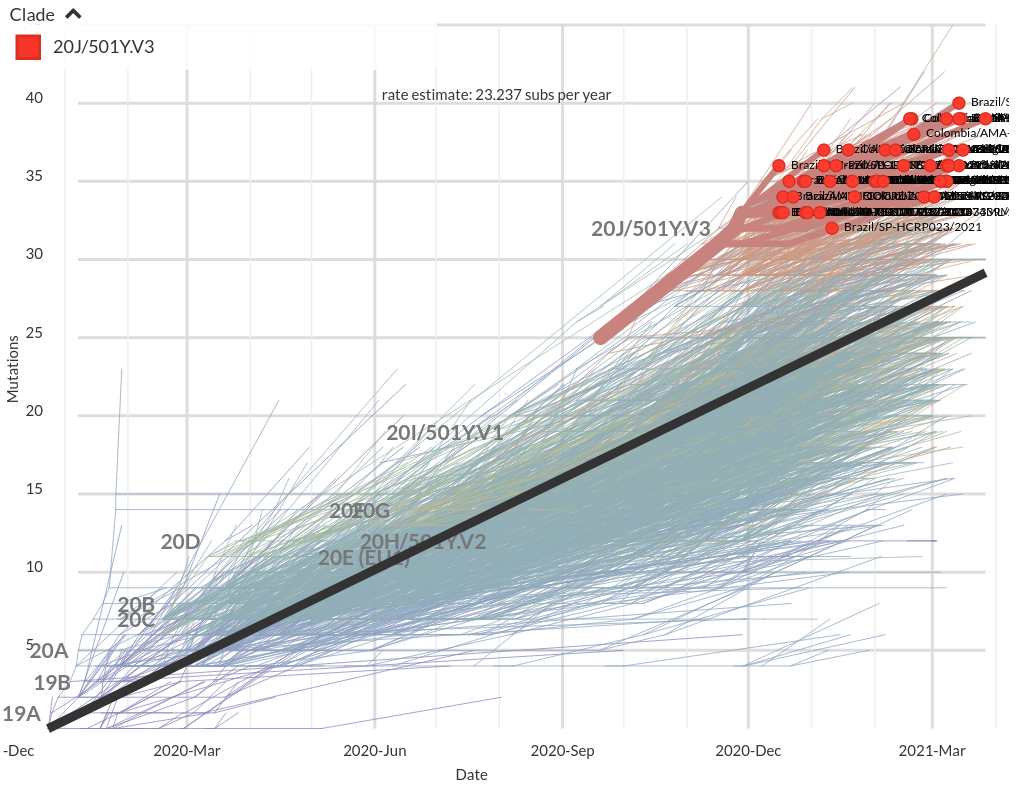
in SARS-CoV-2 a new mutation accumulates about every 2 weeks



- Influenza viruses evolve to avoid human immunity
- Vaccines need frequent updates

Predicting evolution

Given the branching pattern:
- can we predict fitness?
- pick the closest relative of the future?
Fitness inference from trees
$$P(\mathbf{x}|T) = \frac{1}{Z(T)} p_0(x_0) \prod_{i=0}^{n_{int}} g(x_{i_1}, t_{i_1}| x_i, t_i)g(x_{i_2}, t_{i_2}| x_i, t_i)$$
RN, Russell, Shraiman, eLife, 2014
Prediction of the dominating H3N2 influenza strain
- no influenza specific input
- how can the model be improved? (see model by Luksza & Laessig)
- what other context might this apply?
Real-time tracking of SARS-CoV-2
- hundreds of new sequences every day
- more than >6M sequences right now
- comprehensive analysis require hours to days to complete
→ requires continuous analysis and easy dissemination
→ interpretable and intuitive visualization
nextstrain.org
joint project with Trevor Bedford & his lab
Emergence and dominance of VoCs
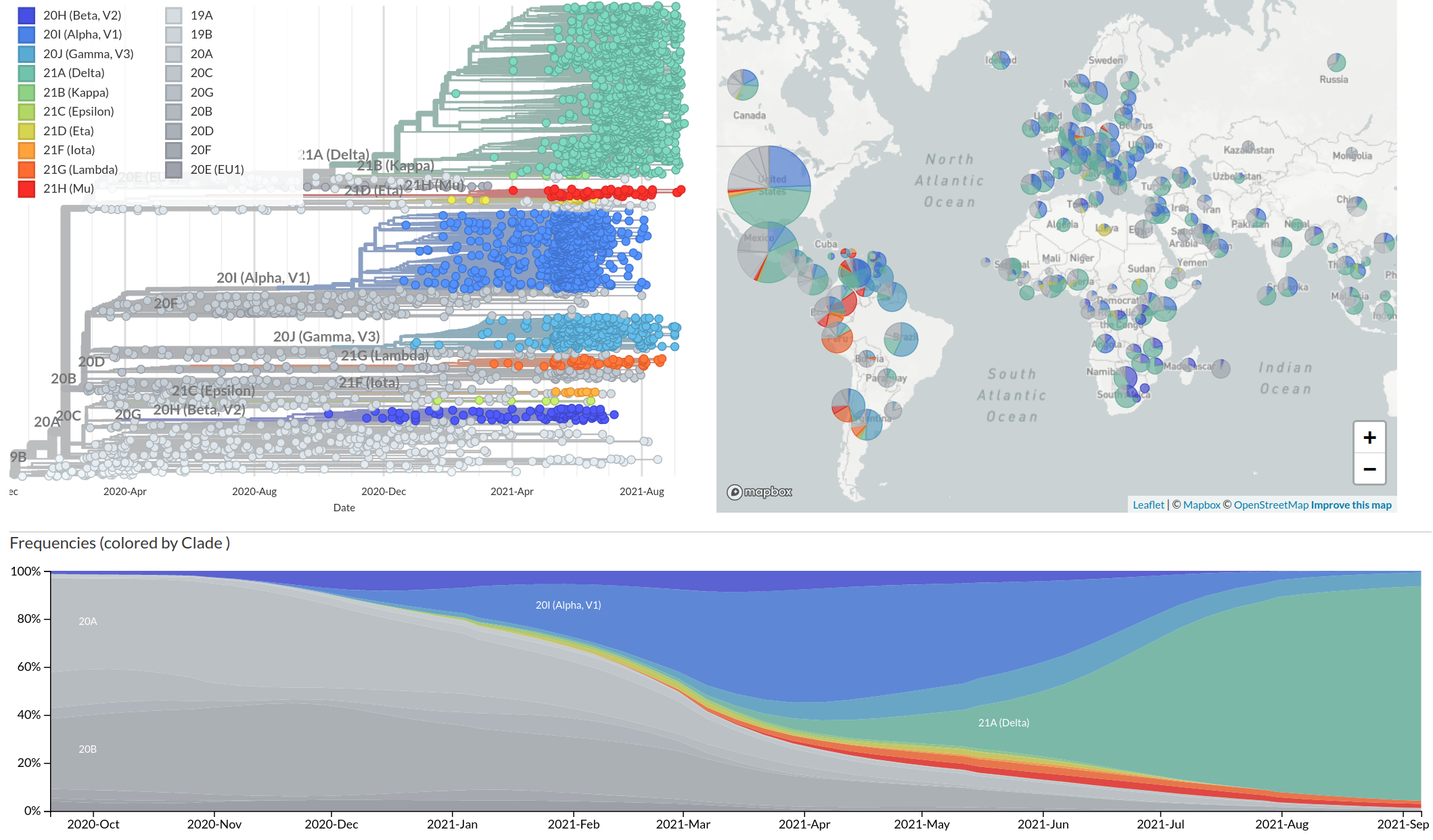
VoCs have more mutations than expected...
 nextstrain
nextstrain
Acknowledgments
- Fabio Zanini
- Jan Albert
- Johanna Brodin
- Christa Lanz
- Göran Bratt
- Lina Thebo
- Vadim Puller



Acknowledgments
- Emma Hodcroft
- Moira Zuber
- Inaki Comas
- Fernando Gonzales
- Tanja Stadler
- Sarah Nadeau
- Tim Vaughan
- Jesse Bloom
- David Veesler
