Reconstructing, tracking, and predicting viral spread and evolution
Richard Neher
Biozentrum & SIB, University of Basel
slides at neherlab.org/202203_Qlife.html
Human seasonal influenza viruses

Positive tests for influenza in the USA by week
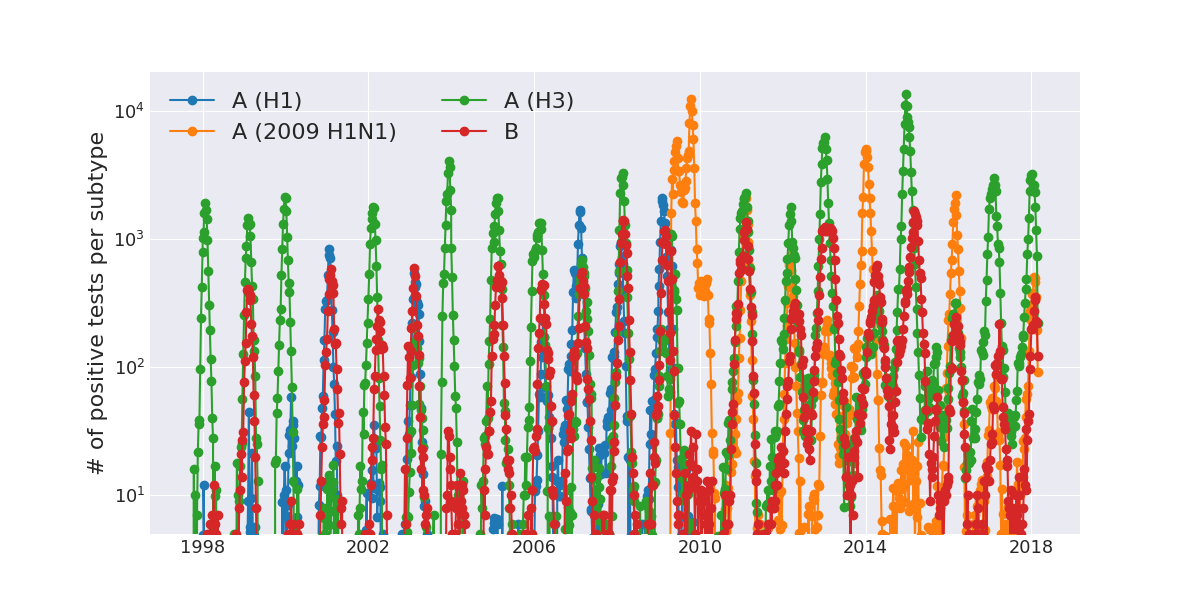 Data by the US CDC
Data by the US CDC
Human corona viruses have pronounced seasonal prevalence (Sweden)
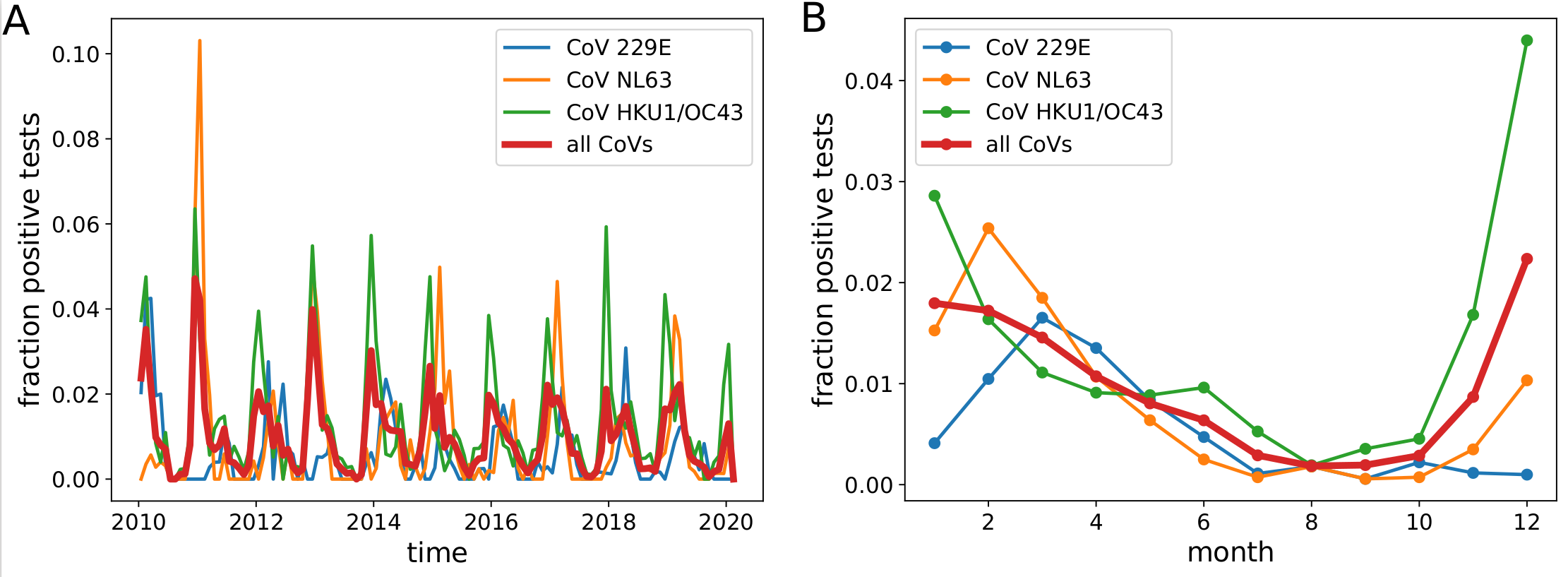
- Respiratory virus incl seasonal CoVs show seasonality
- Forcing through human behavior (indoor/outdoor activities).
- Absolute effect of seasonality unknown
- The two alpha and beta coronaviruses alternate
Viral dynamics beyond case counts?
Sequences record the spread of pathogens




Mutations accumulate at a rate of $10^{-5}$ per site and day!
Genomic analysis to reconstruct pathogen spread and evolution
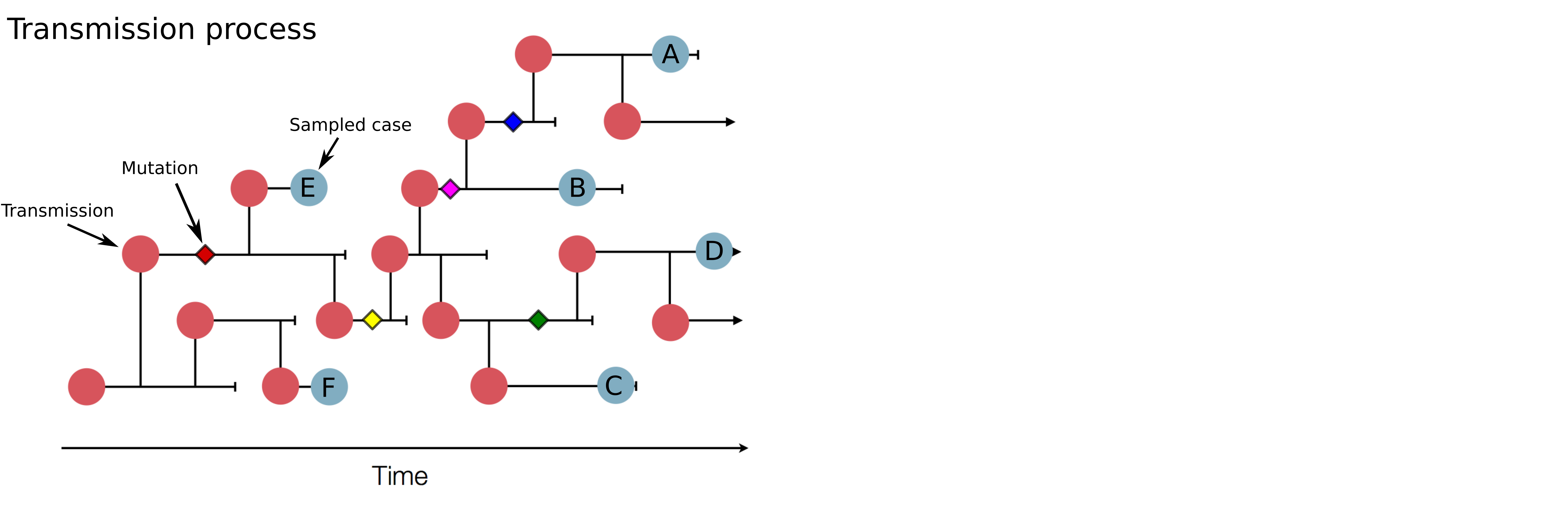
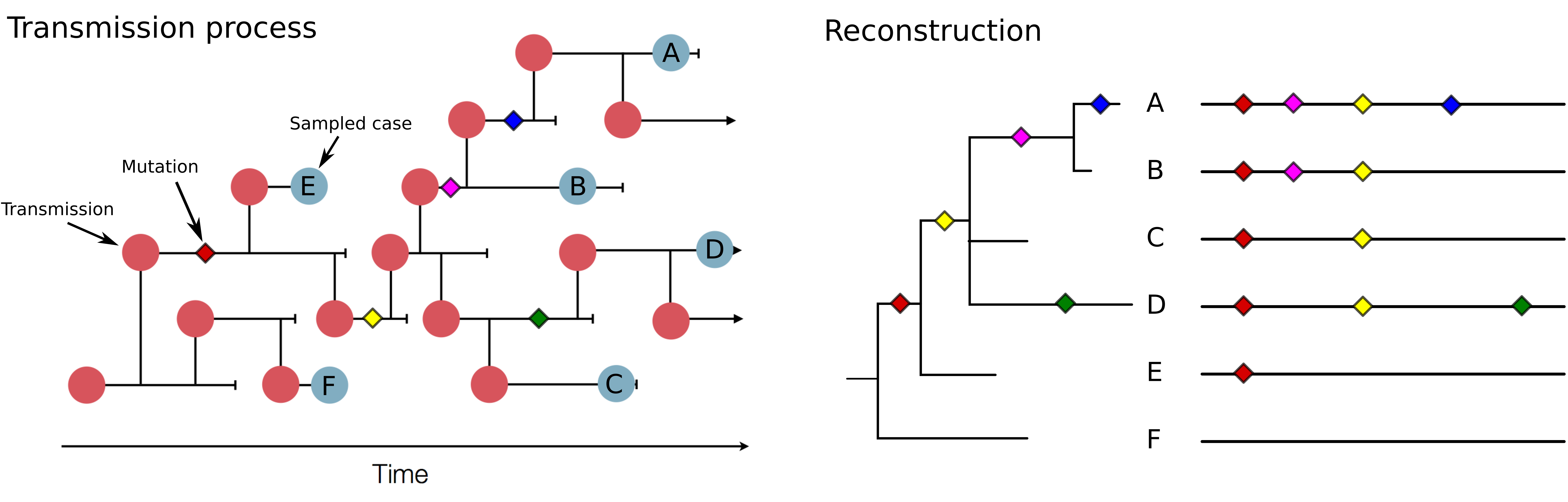
Mutations allow to reconstruct how pathogens spread -- clusters, introductions, etc
Mutations can result in phenotypic change -- immune escape, drug resistance, host adaptation
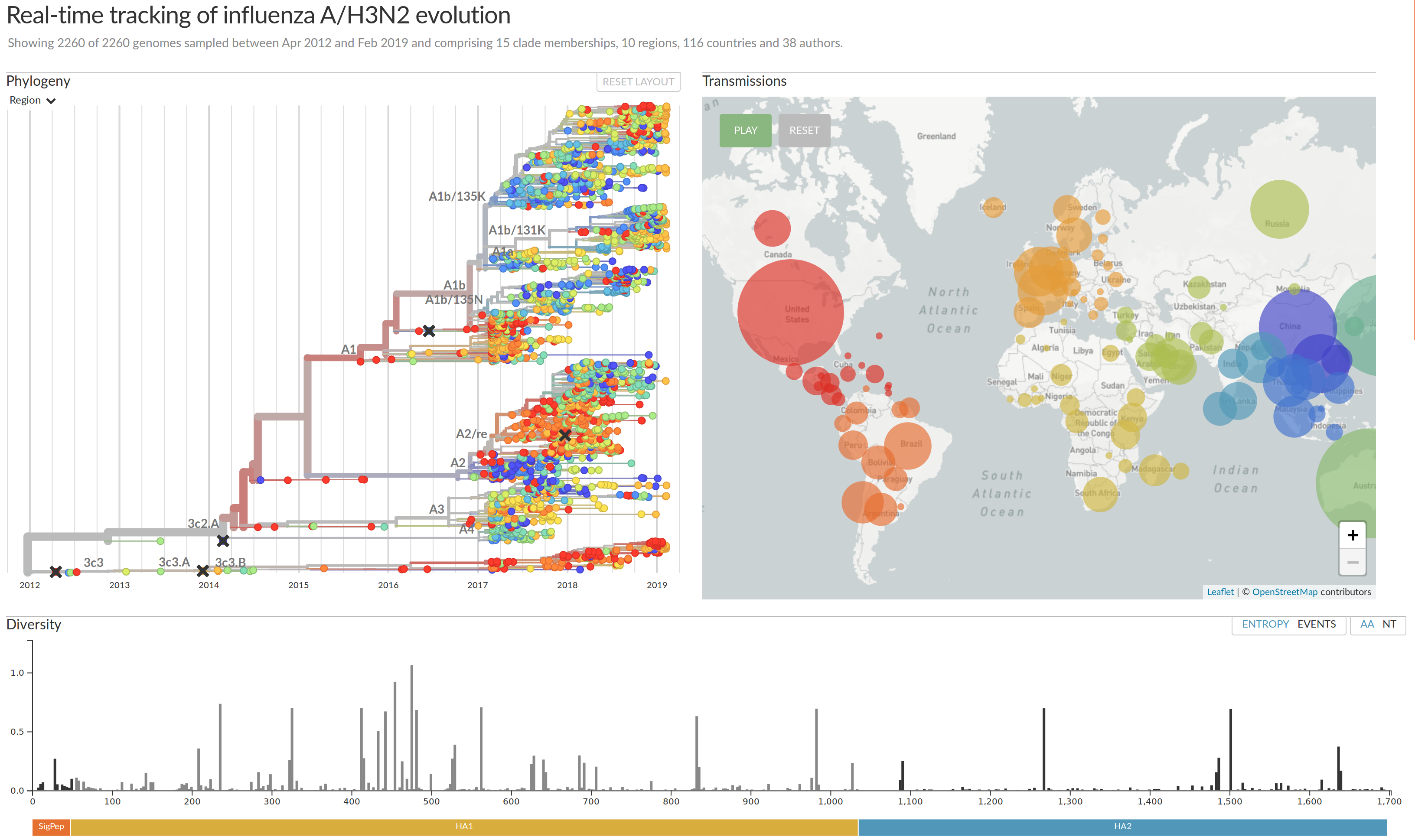
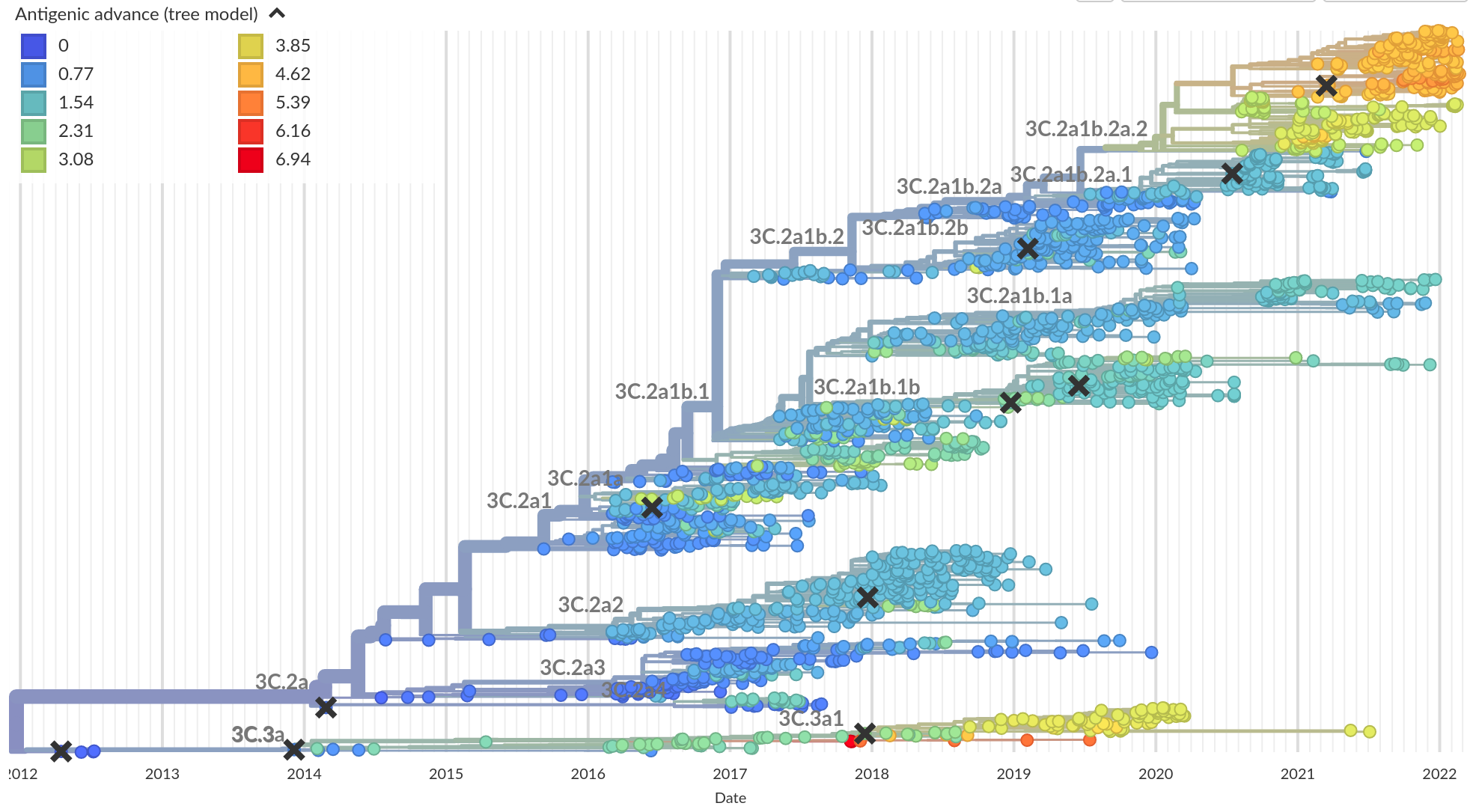
Influenza A/H3N2



- Influenza viruses evolve to avoid human immunity
- Vaccines need frequent updates

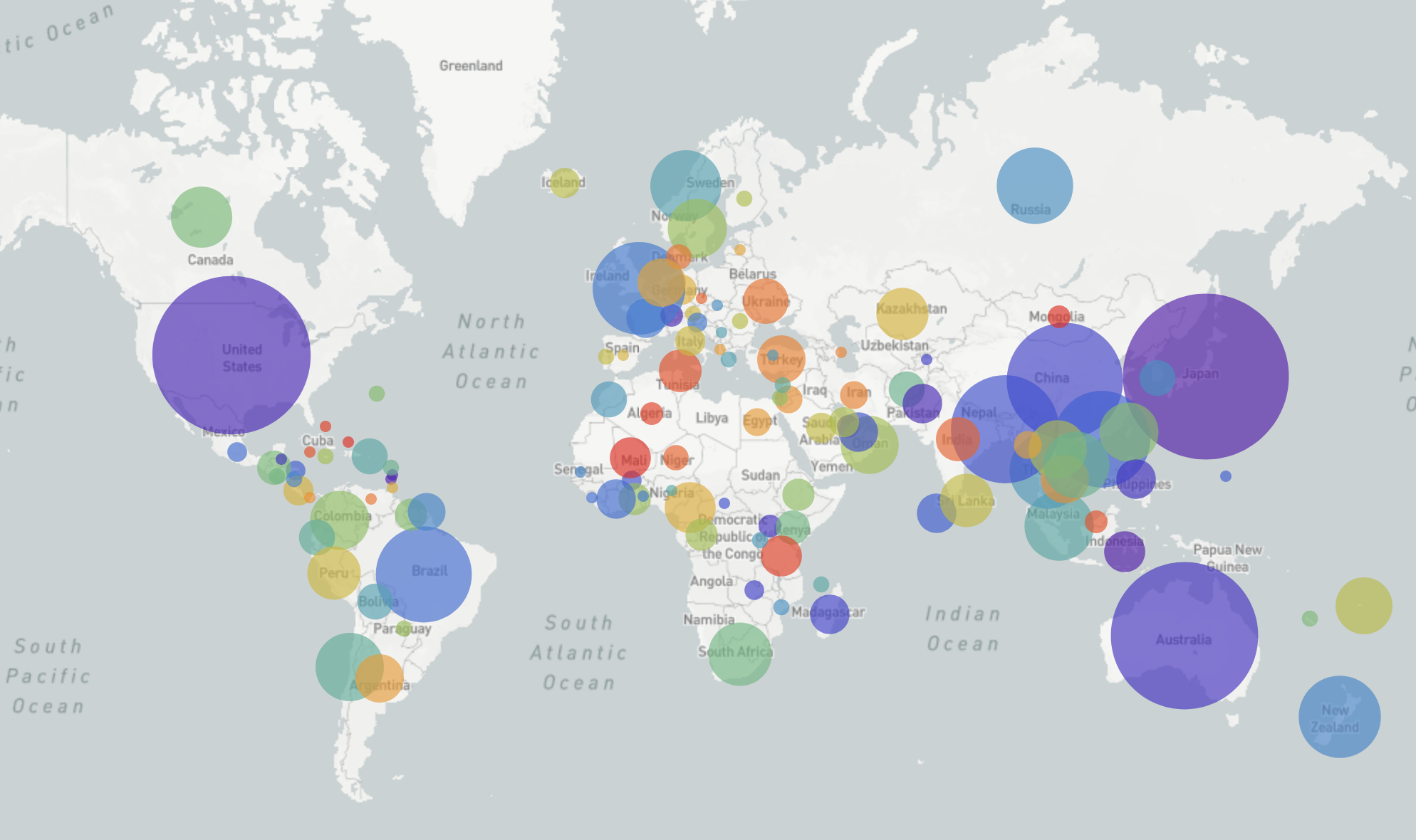
nextflu.org
joint project with Trevor Bedford & his lab
Beyond tracking: can we predict?
Prediction of the dominating H3N2 influenza strain
- Explicit fitness scores based on specific mutations
→ mutations in previously characterized epitopes
→ mutations that likely reduce fitness
→ mostly historically ascertained - Phylogenetic indicators to spot rapidly expanding clades
- Laboratory data (antigenicity, virulence)
- Human serology
Mutational signatures -- epitope mutations
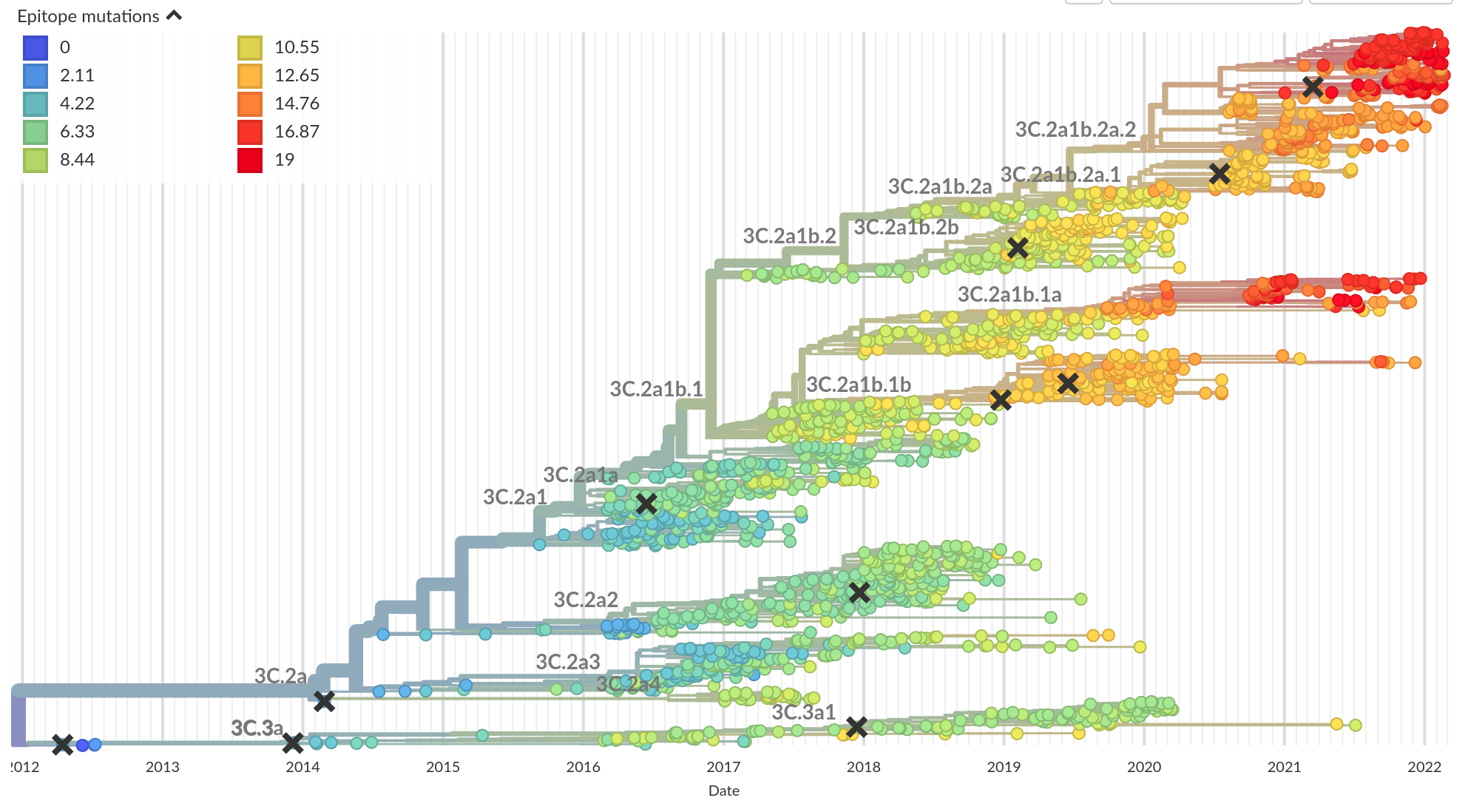 Luksza and Laessig, Nature, 2014
Luksza and Laessig, Nature, 2014
Phylogenetic indicators -- LBI
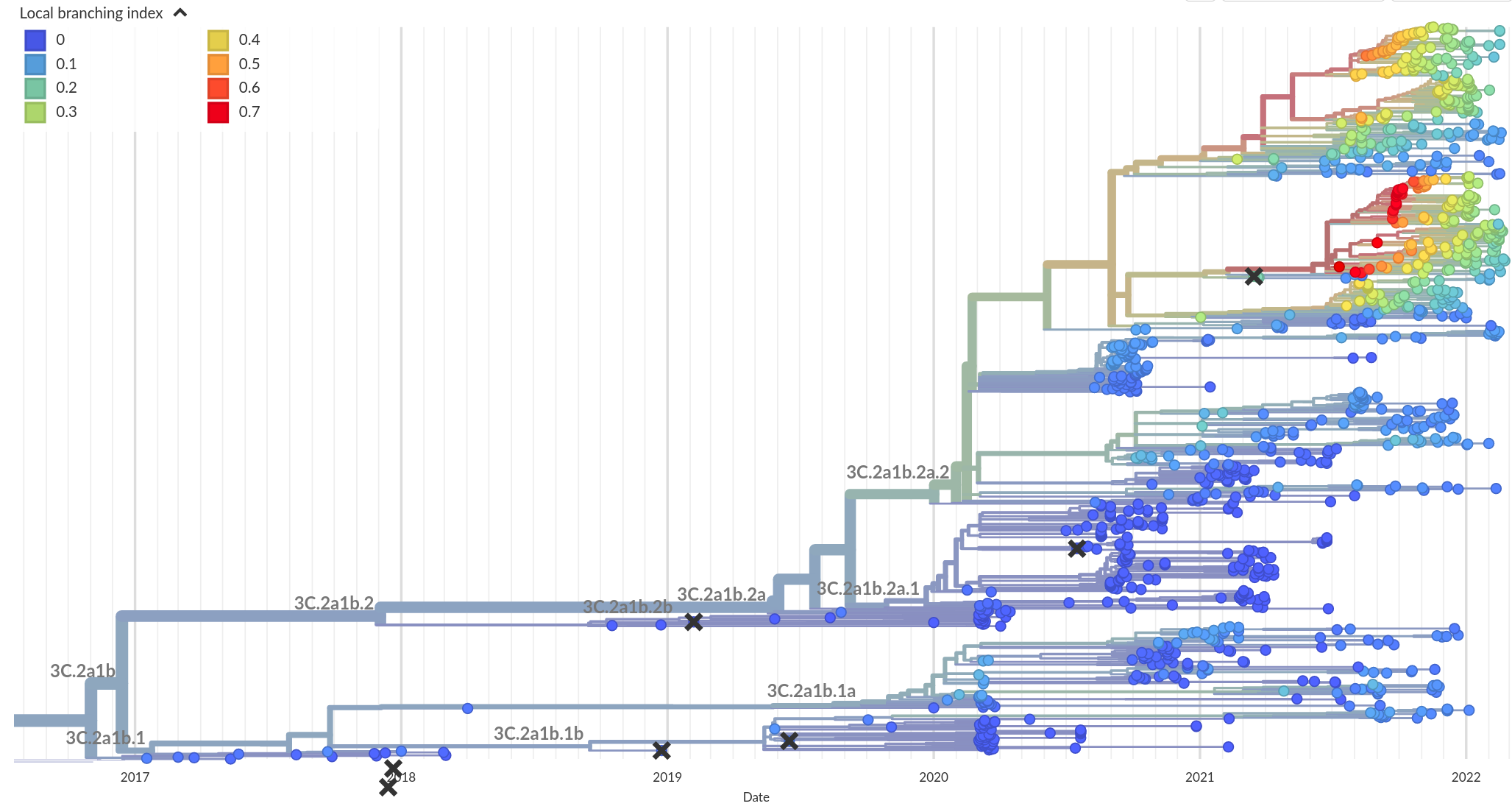 RN, Russell, Shraiman, eLife, 2014
RN, Russell, Shraiman, eLife, 2014
Hemagglutination Inhibition assays
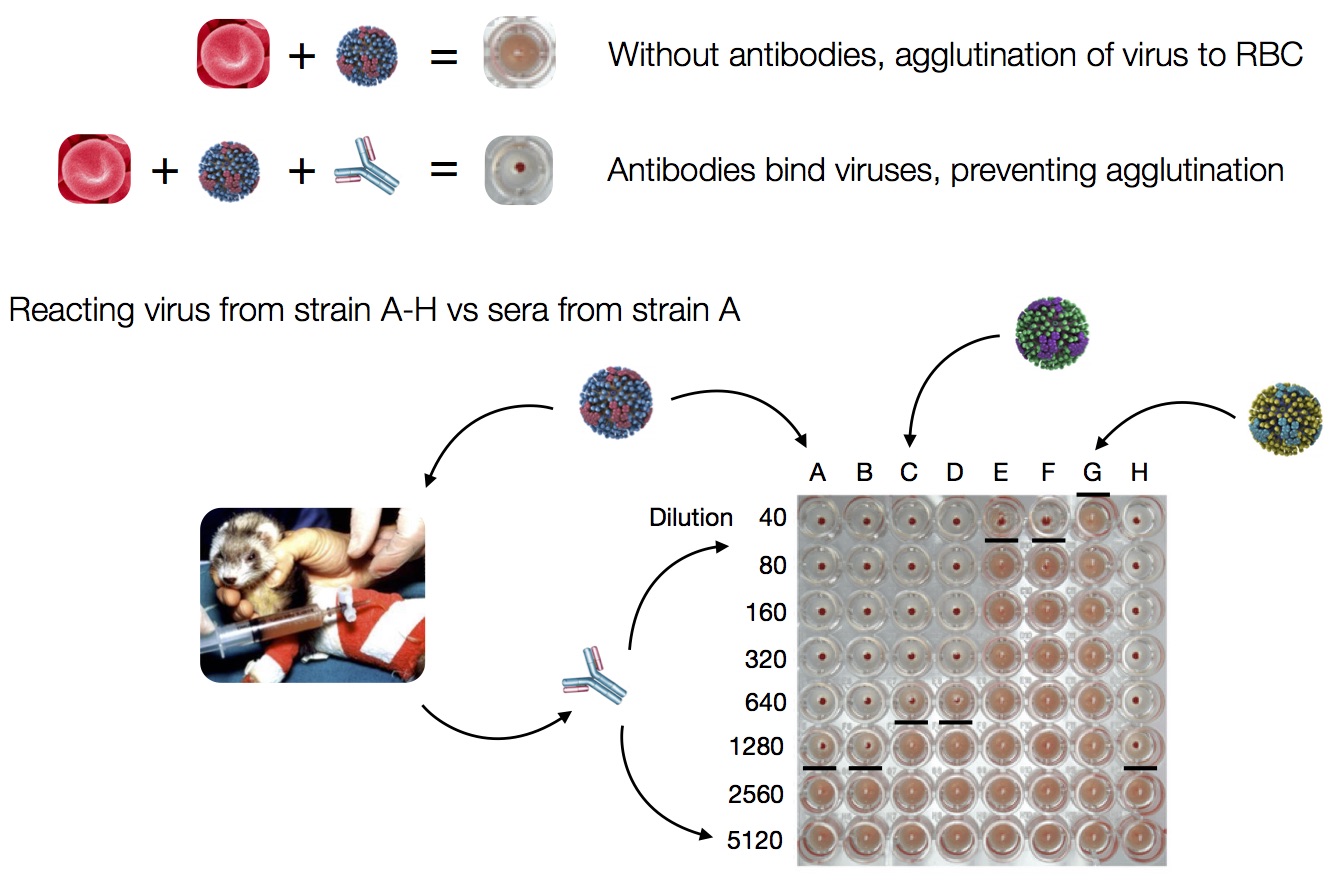
Antigenic distance tables
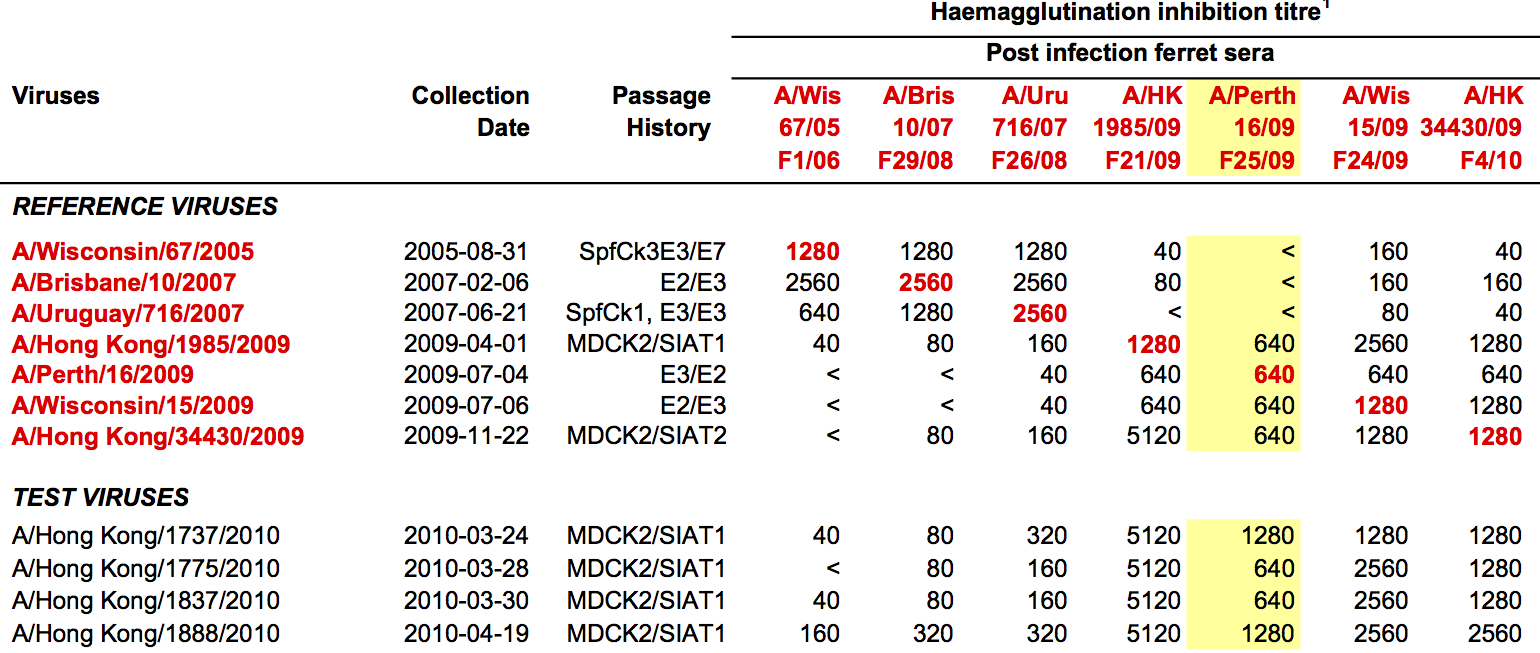
- Long list of distances between sera and viruses
- Tables are sparse, only close by pairs
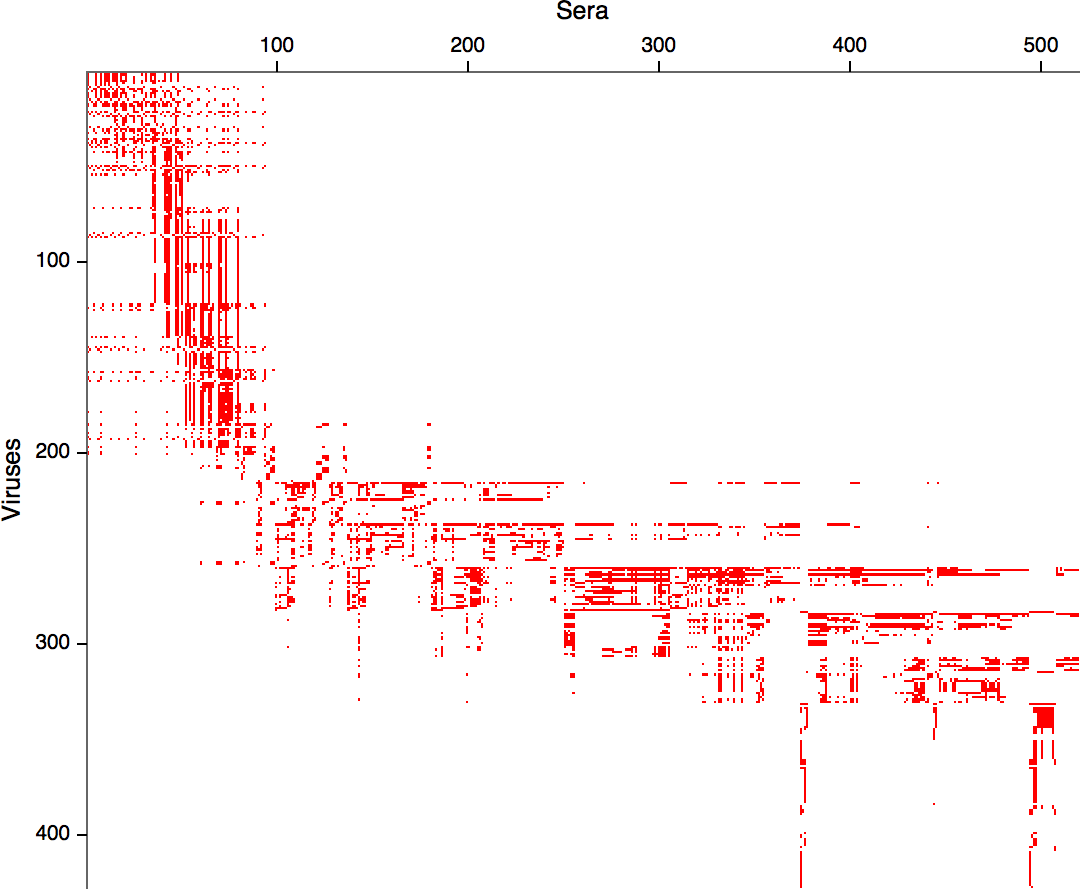
Integrating antigenic and molecular evolution -- ferret serology
 RN et al, PNAS, 2017
RN et al, PNAS, 2017
Accurate prediction remains a challenge!
SARS-CoV-2
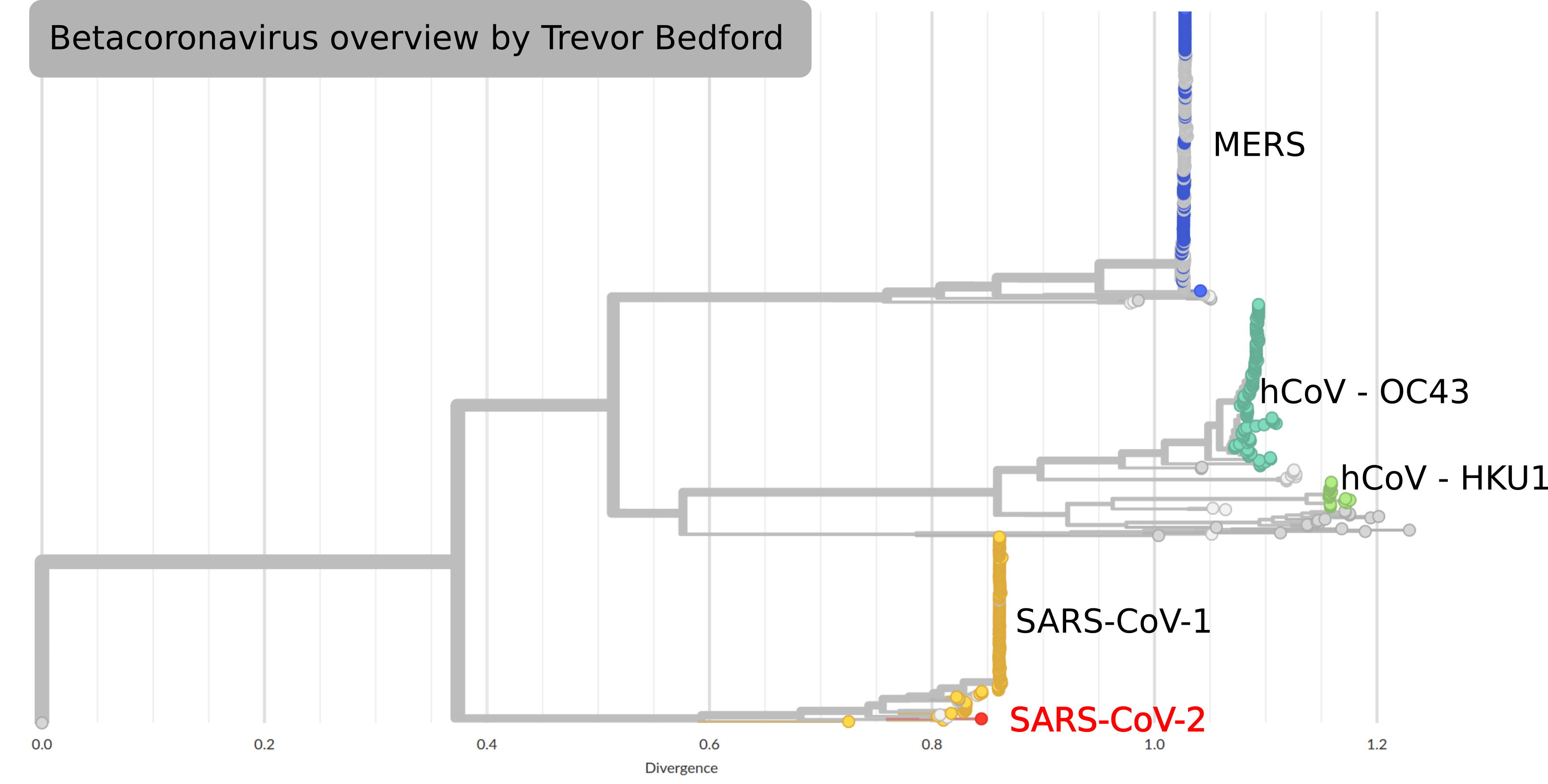 by Trevor Bedford
by Trevor Bedford
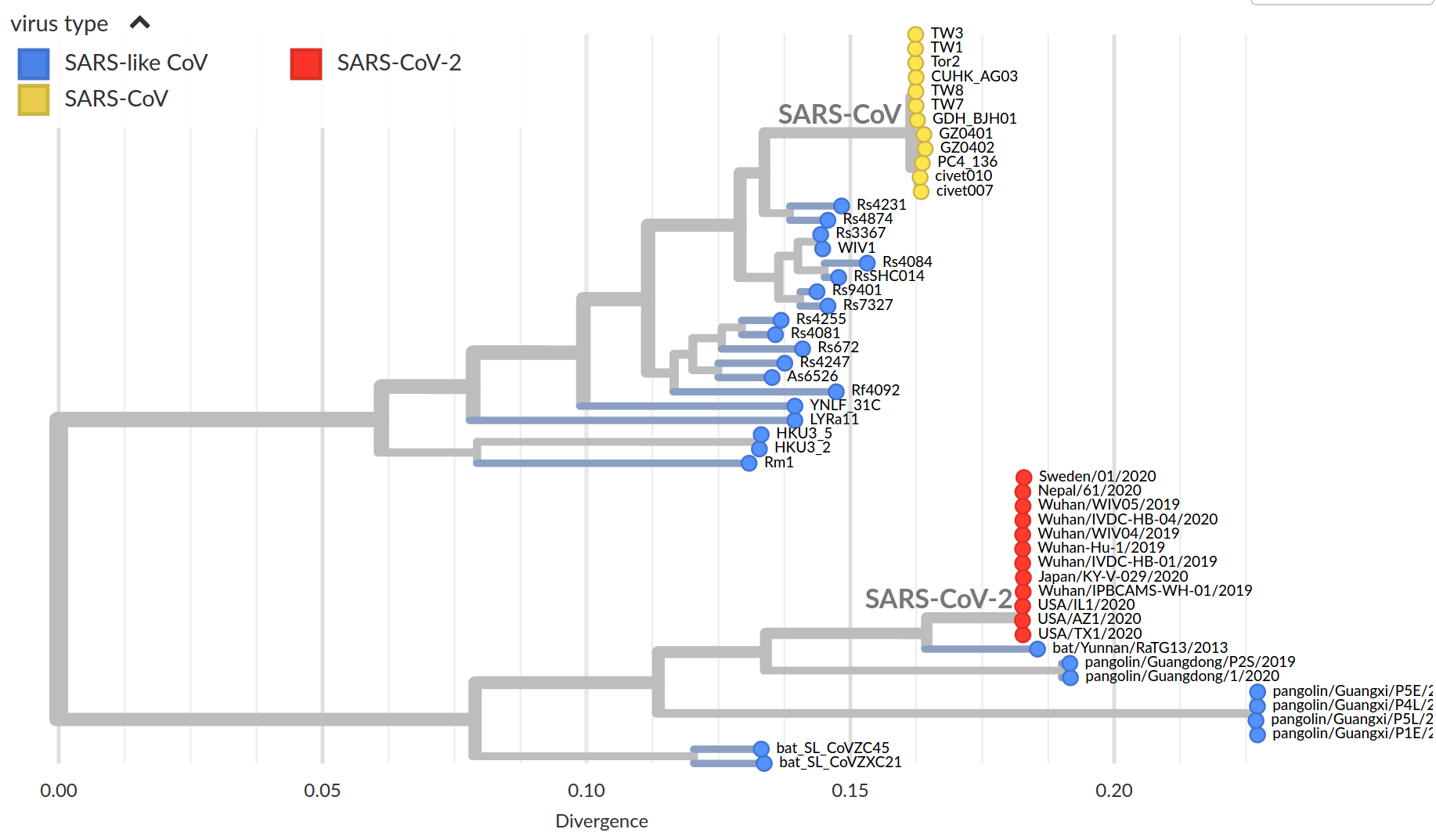 by Trevor Bedford
by Trevor Bedford
Available data on Jan 26 2020
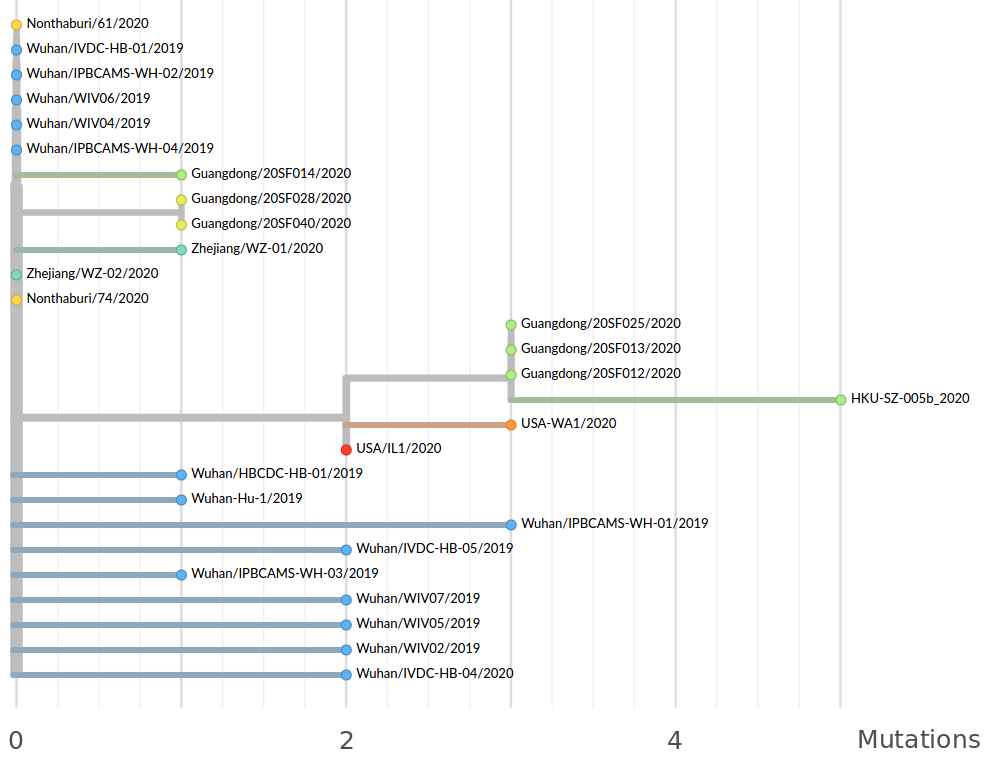
Early genomes differed by only a few mutations, suggesting very recent emergence
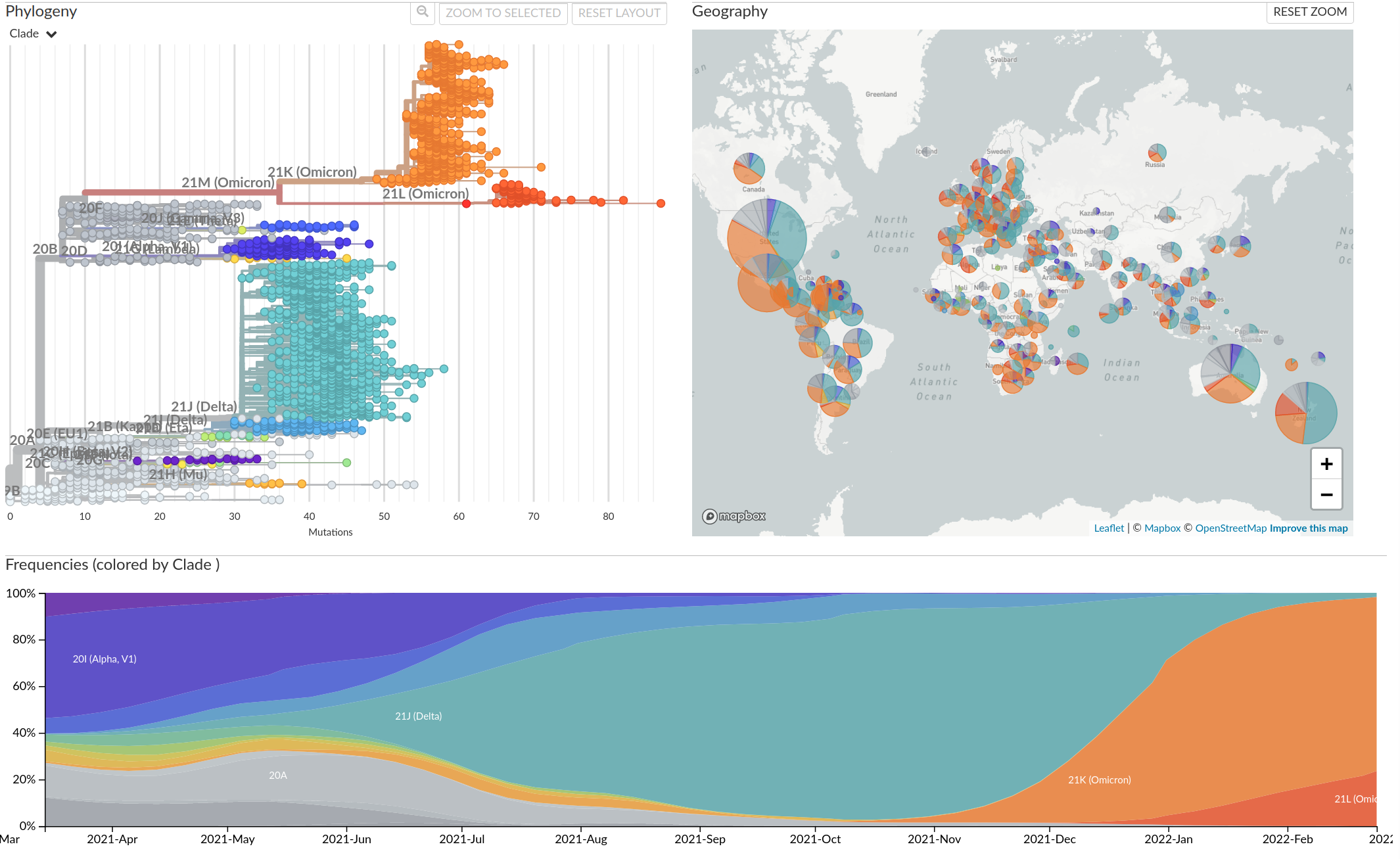
Tracking diversity and spread of SARS-CoV-2 in Nextstrain
Spike mutations in Omicron
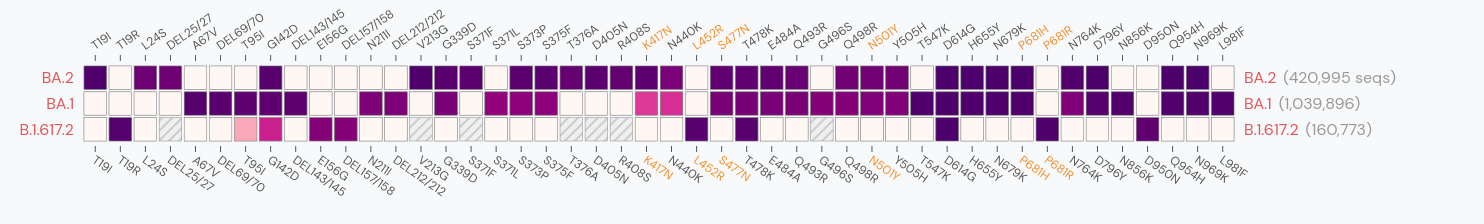
Prediction of SARS-CoV-2 evolution
- Fitting frequency dynamics to mutations and sequence/variant features.
→ complex epistatic interactions prevent extrapolation to new variants - Using laboratory data to map effects of all possible mutations (deep mutational scanning).
→ immune escape can be assessed from sequence, virulence and fitness not
Too many aspects of SARS-CoV-2 evolution are unknown to make meaningful predictions.
Medium term dynamics of SARS-CoV-2 is very uncertain
- Will we start seeing second and third generation variants, as opposed to sister variants?
- Will we the saltatory dynamics with heavily diverged variants continue?
- Will a more diverse immunity landscape slow down future variant dynamics?
- Will waning/antigenic evolution slow down and give rise to annual or even rarer waves?
So far, independent variants have dominated successively

Going forward, how diverse is SARS-CoV-2 going to be?
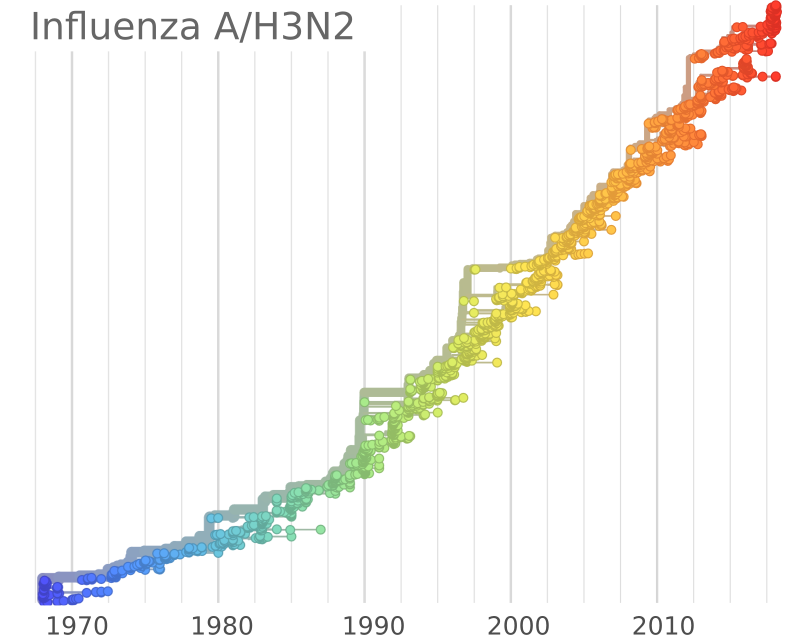
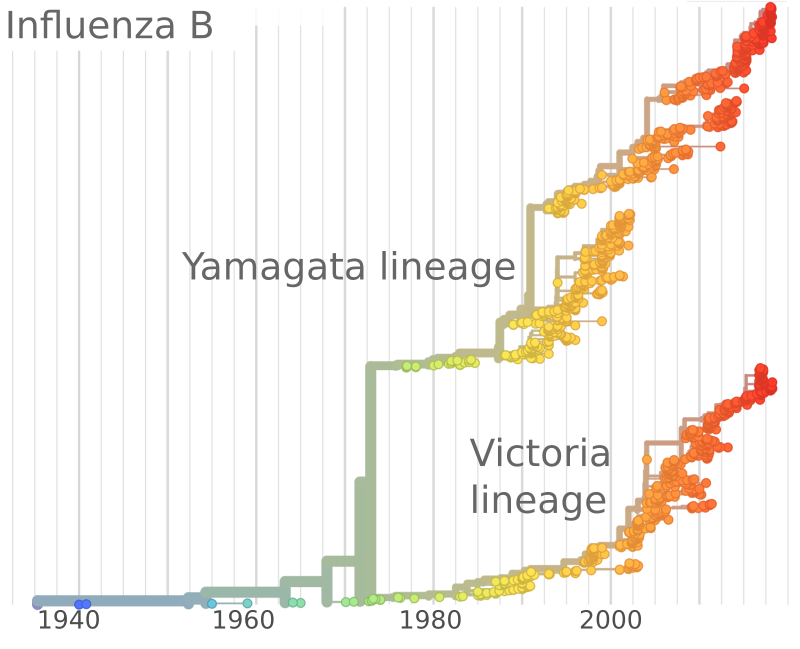
Influenza and Theory acknowledgments





- Boris Shraiman
- Colin Russell
- Trevor Bedford
- Pierre Barrat
- Oskar Hallatschek
- All the NICs and WHO CCs that provide influenza sequence data
- The WHO CCs in London and Atlanta for providing titer data



Acknowledgments
Trevor Bedford and his lab -- terrific collaboration since 2014

especially James Hadfield, Emma Hodcroft, Ivan Aksamentov, Cornelius Roemer, Moira Zuber, and John Huddleston
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID

