Reconstructing, tracking, and predicting viral spread and evolution
Richard Neher
Biozentrum & SIB, University of Basel
slides at neherlab.org/202205_MPGHD.html
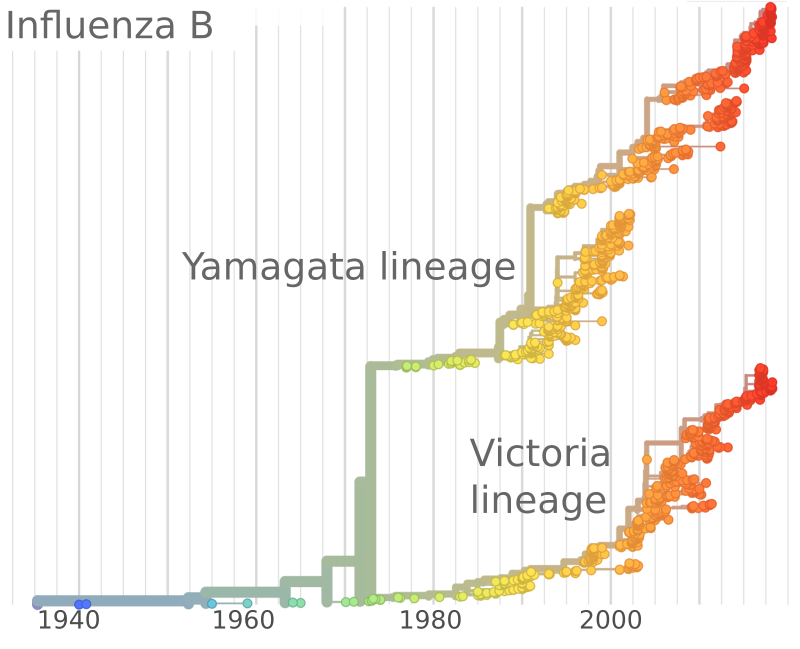
Human seasonal influenza viruses

Positive tests for influenza in the USA by week
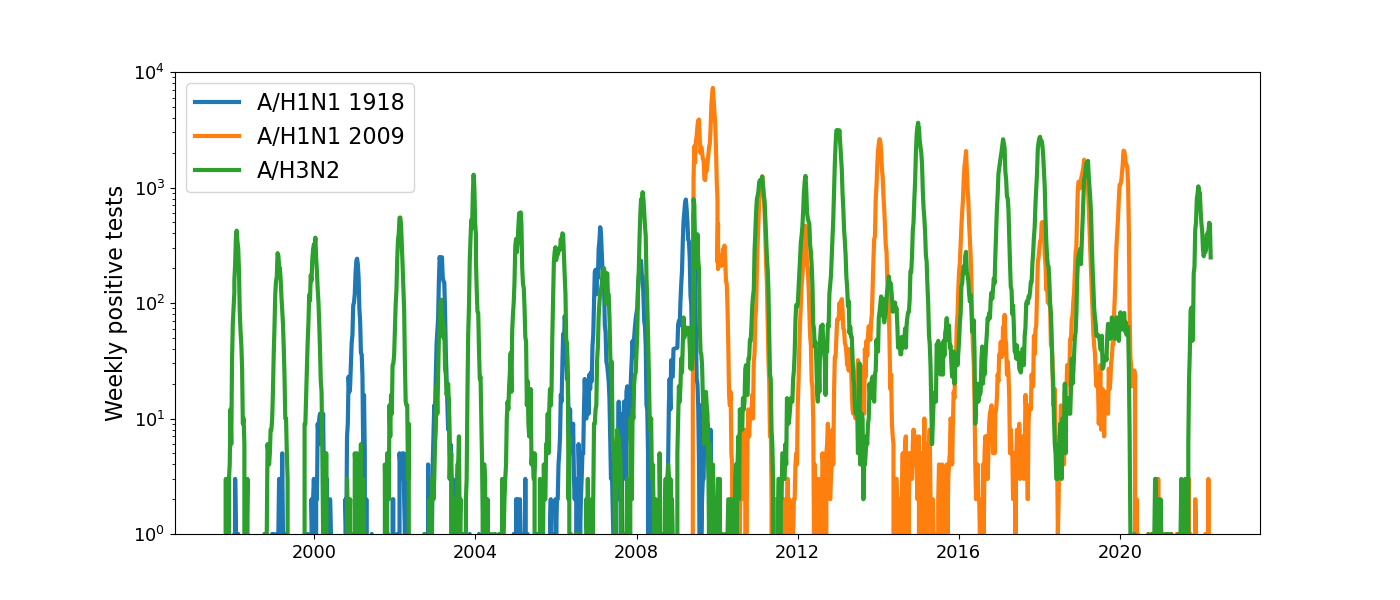 Data by the US CDC
Data by the US CDC
Genomic analysis to reconstruct pathogen spread and evolution
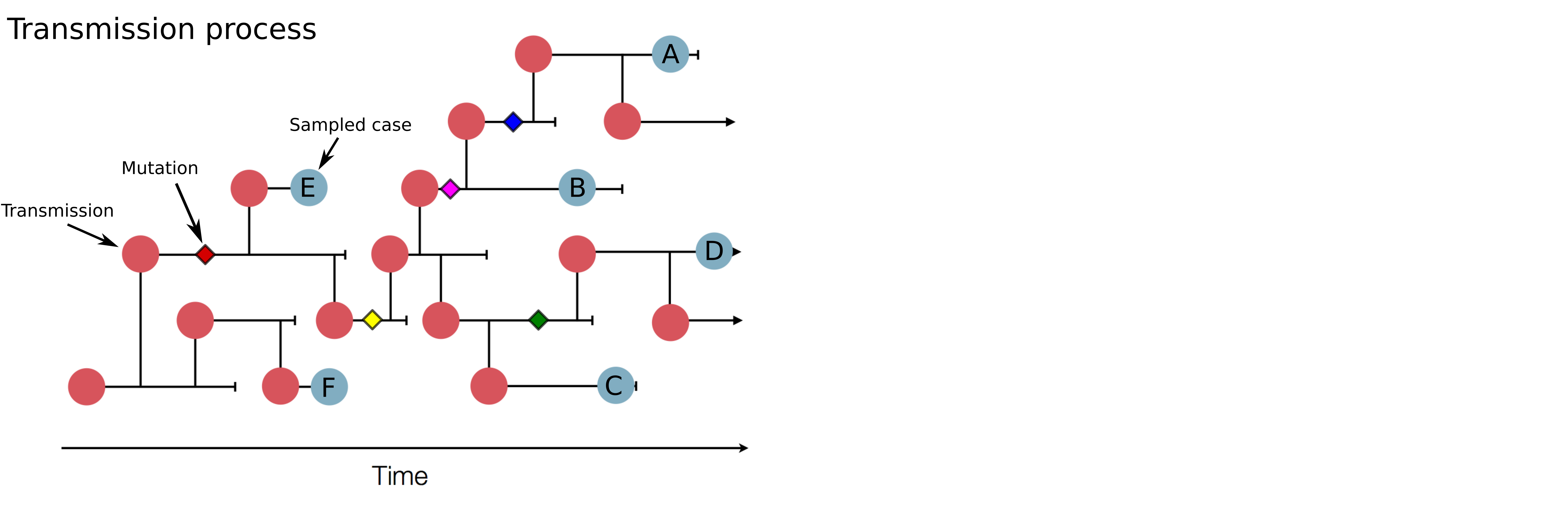
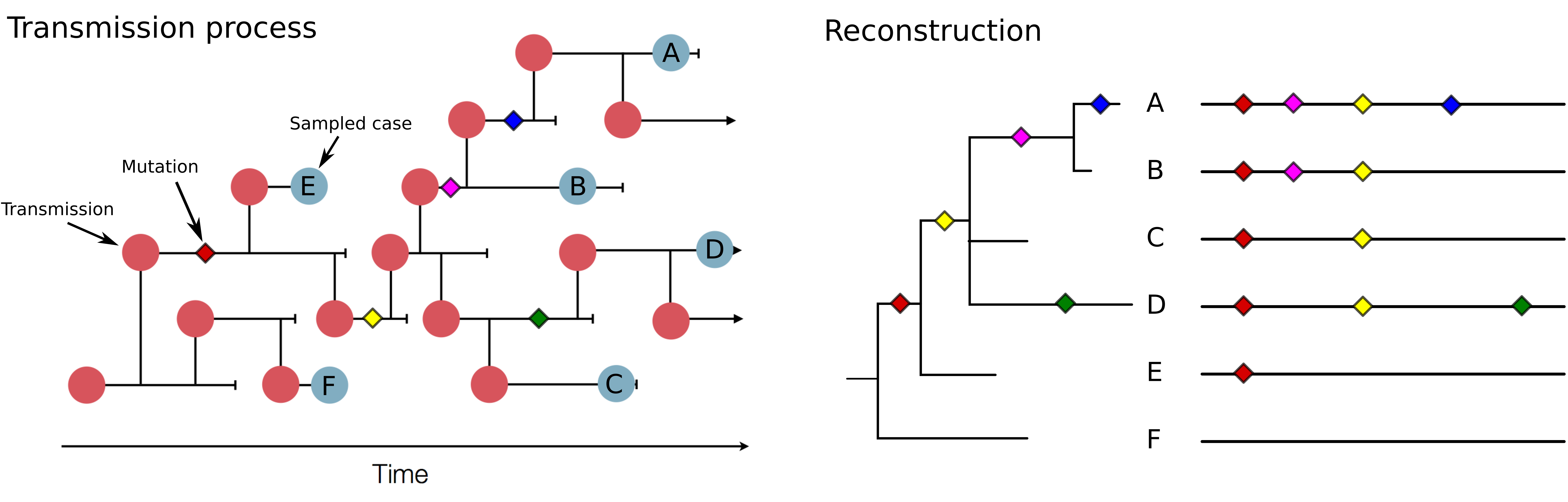
Track how pathogens spread: clusters, introductions, etc
Link genotypic and phenotypic changes: immune escape, drug resistance, host adaptation
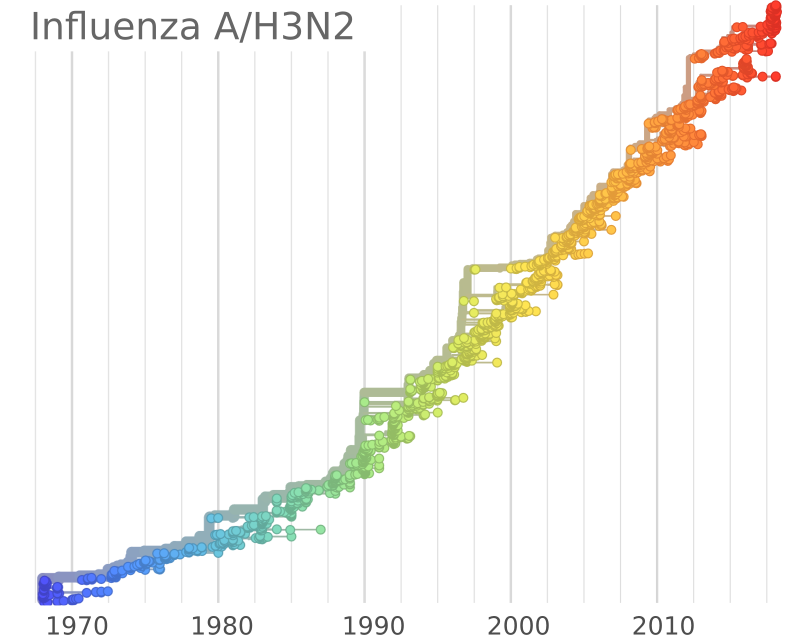
Influenza A/H3N2



- Influenza viruses evolve to avoid human immunity
- Vaccines need frequent updates

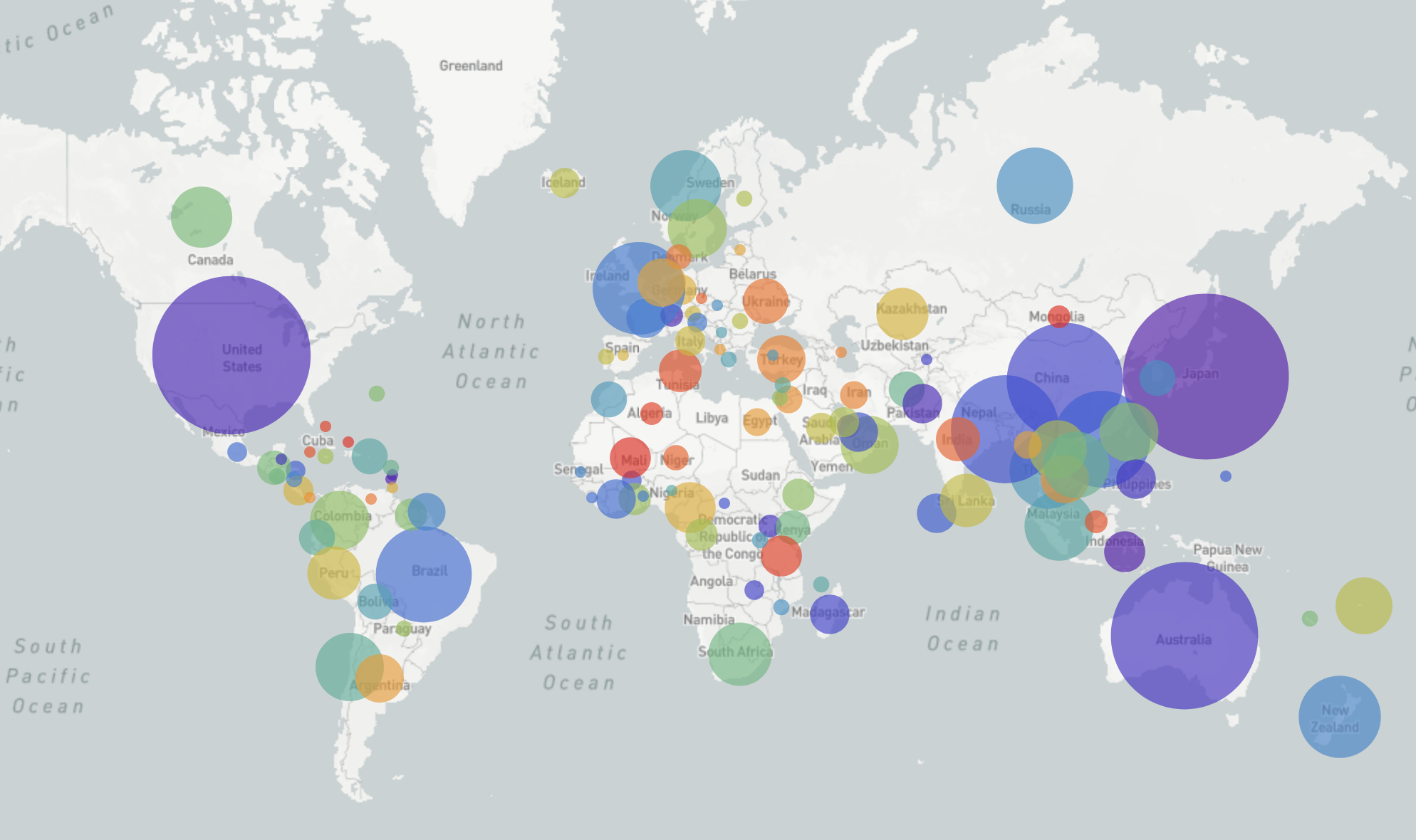
nextflu.org
joint project with Trevor Bedford & his lab
Hemagglutination Inhibition assays
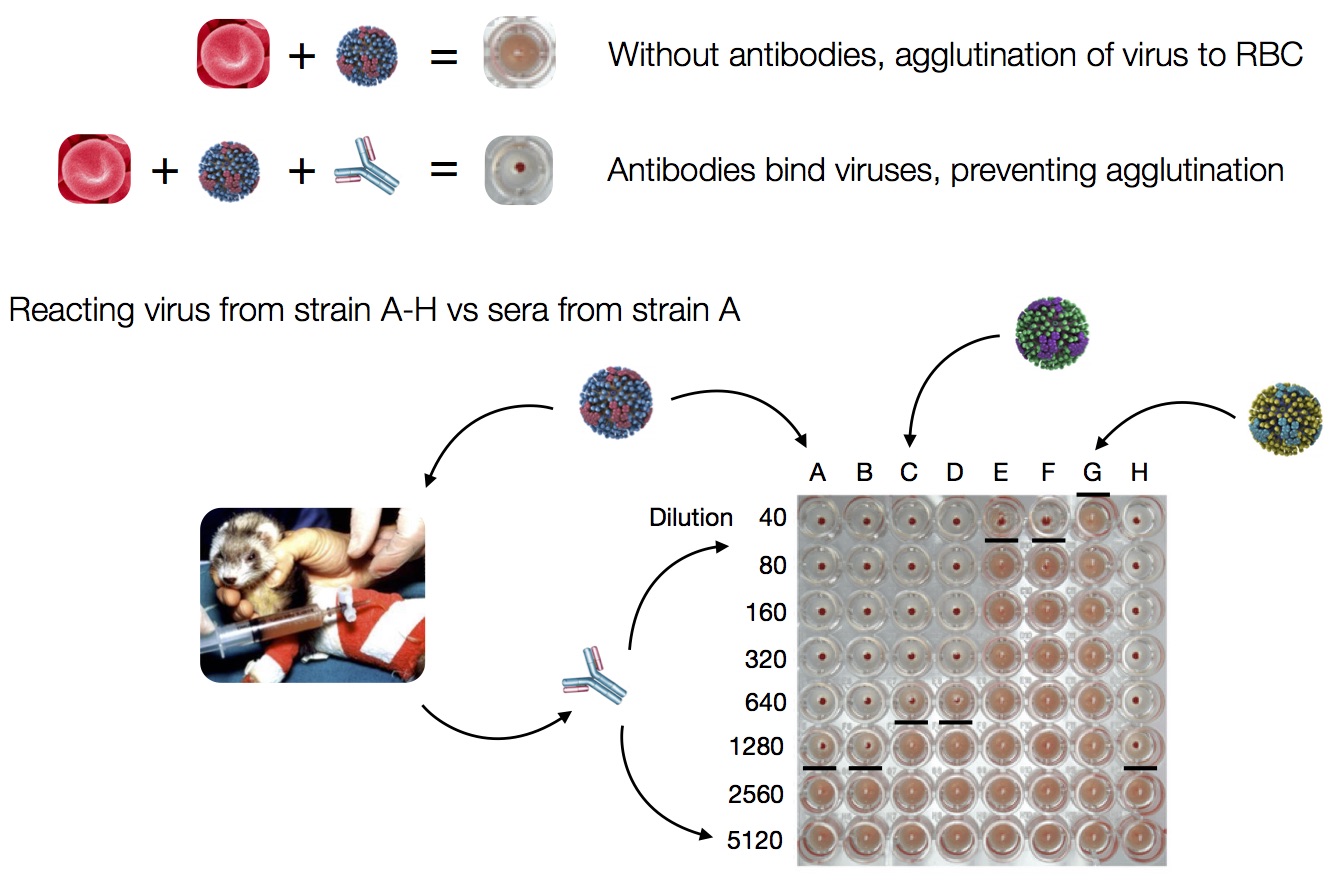
Antigenic distance tables
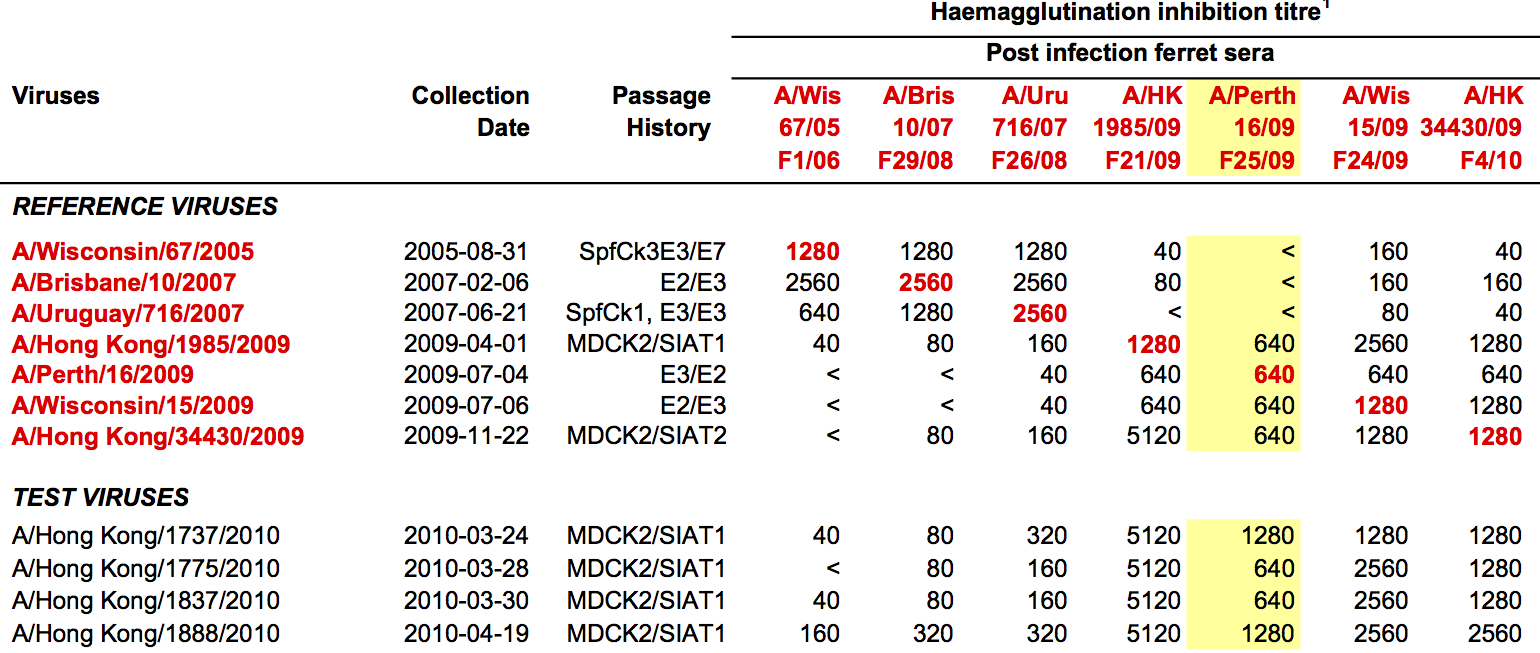
- Long list of distances between sera and viruses
- Tables are sparse, only close by pairs
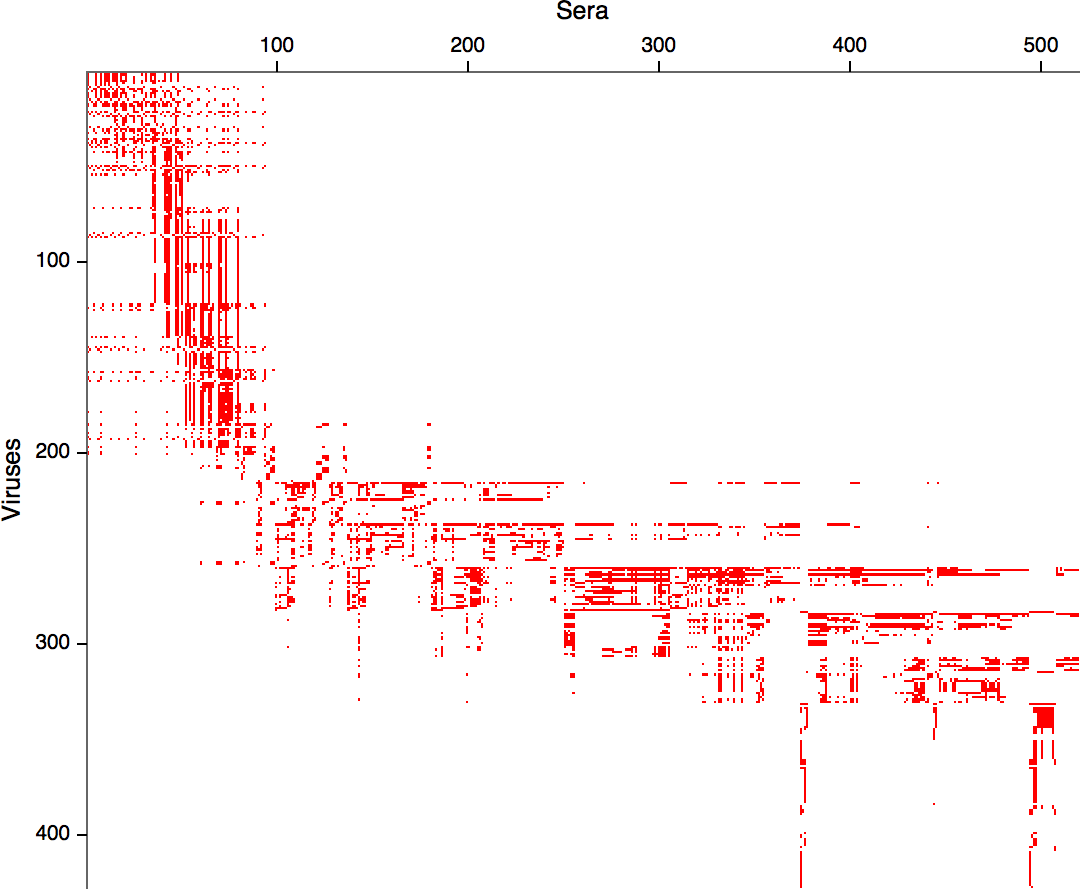
Integrating antigenic and molecular evolution -- ferret serology
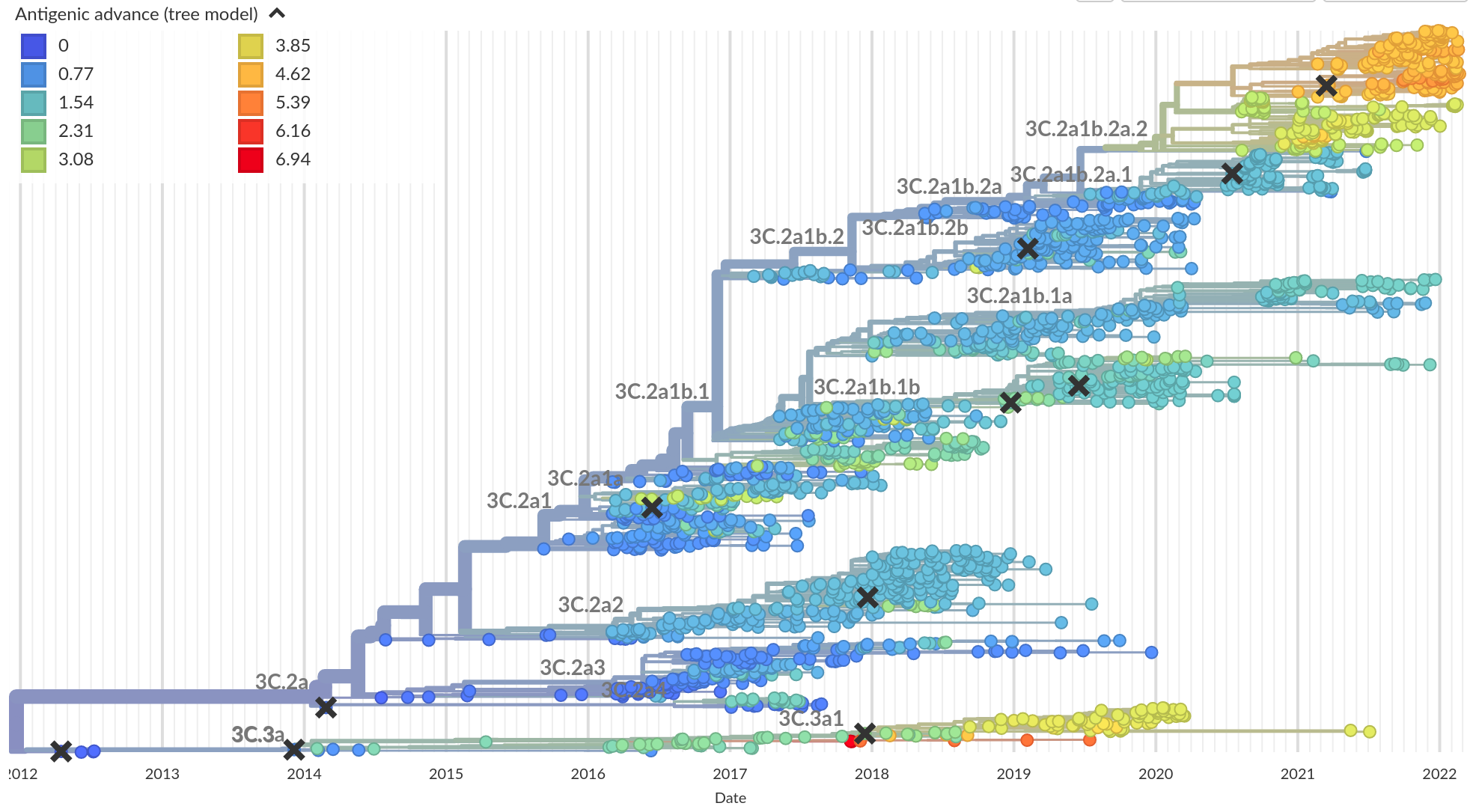 RN et al, PNAS, 2017
RN et al, PNAS, 2017
Integrating antigenic and molecular evolution
- each branch contributes $d_i$ to antigenic distance
- sparse solution for $d_i$ through $l_1$ regularization
Beyond tracking: can we predict?
Prediction of the dominating H3N2 influenza strain
- Explicit fitness scores based on specific mutations
→ mutations in previously characterized epitopes
→ mutations that likely reduce fitness
→ mostly historically ascertained - Phylogenetic indicators to spot rapidly expanding clades
- Laboratory data (antigenicity, virulence)
Prediction of the dominating H3N2 influenza strain
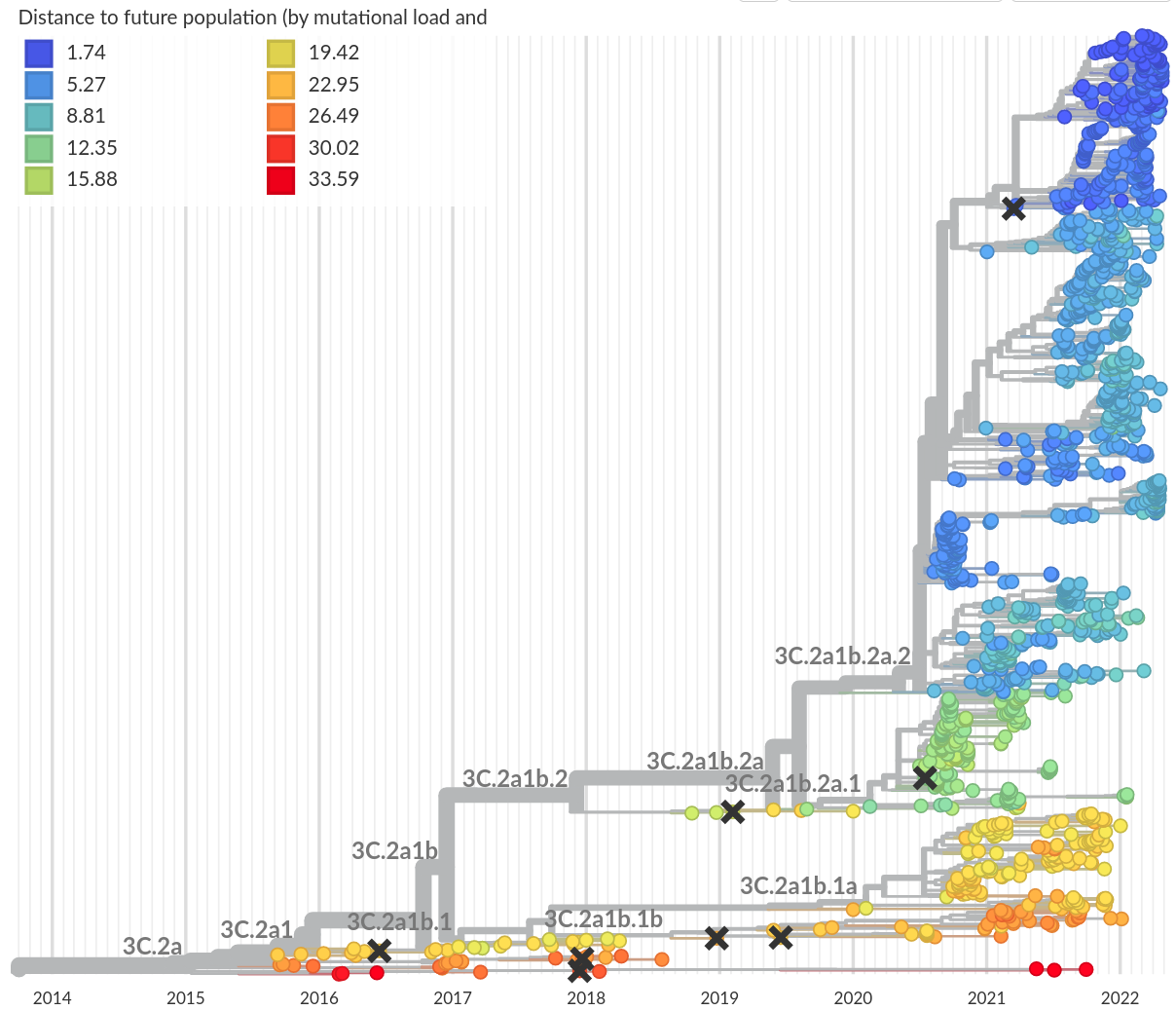
Predicted "distance to future"
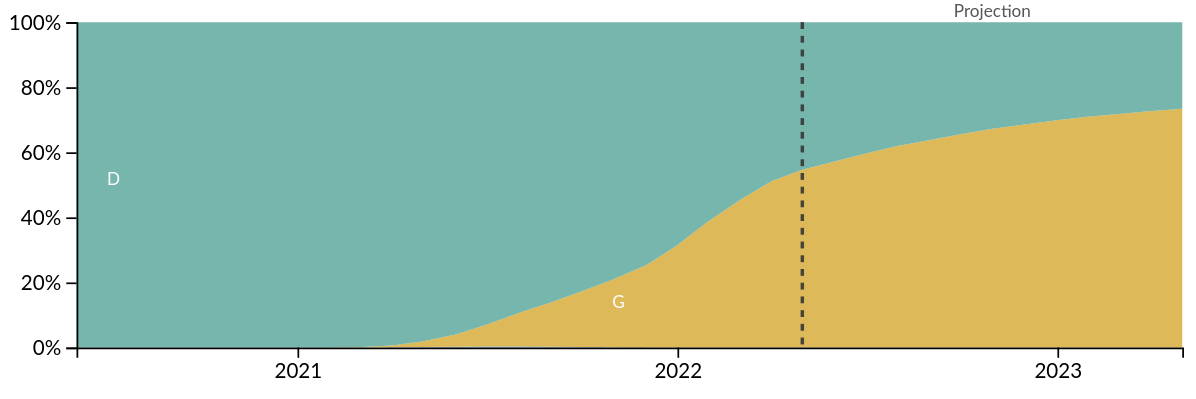
Projection for HA1 mutation 104
SARS-CoV-2
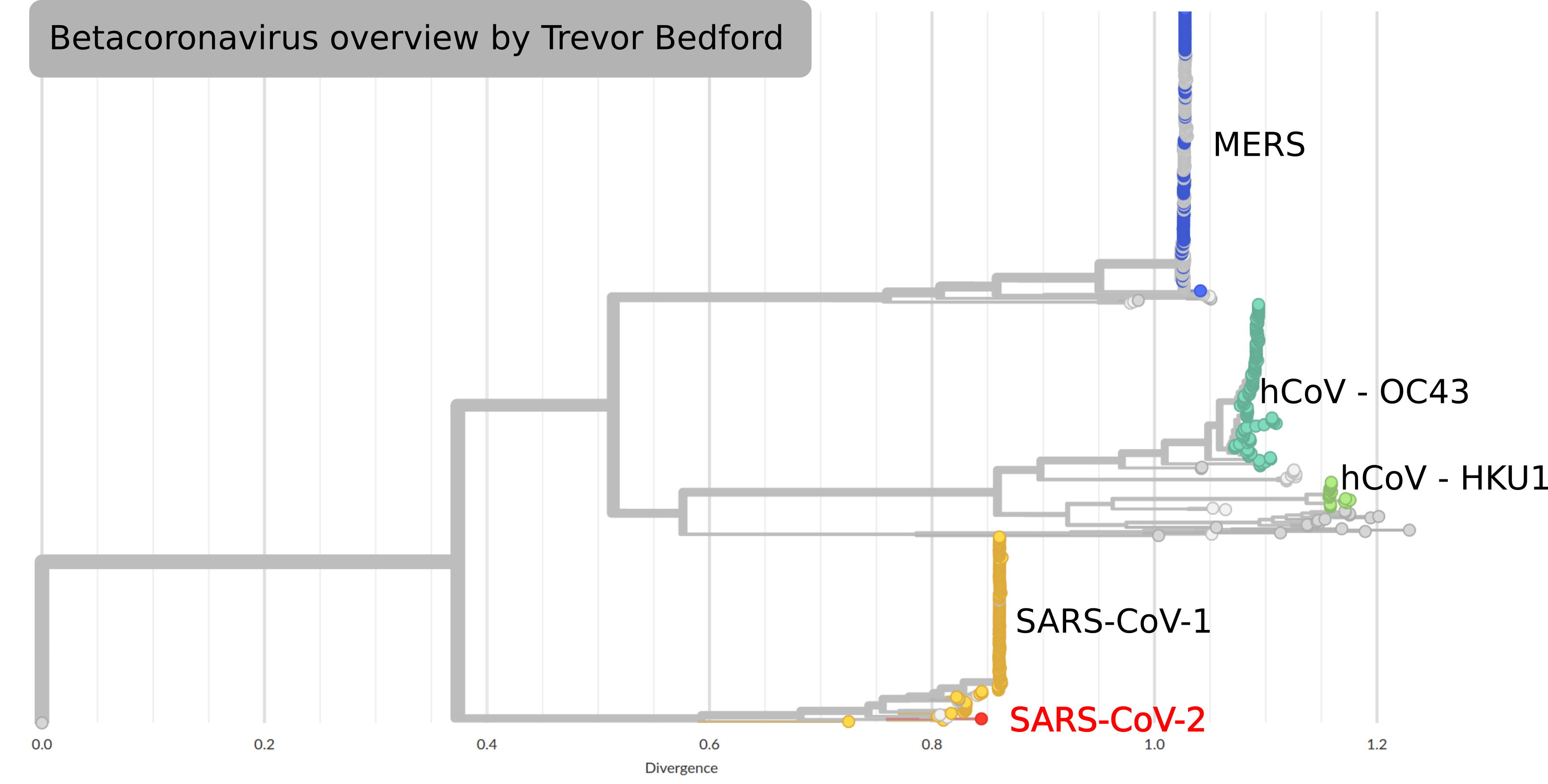 by Trevor Bedford
by Trevor Bedford
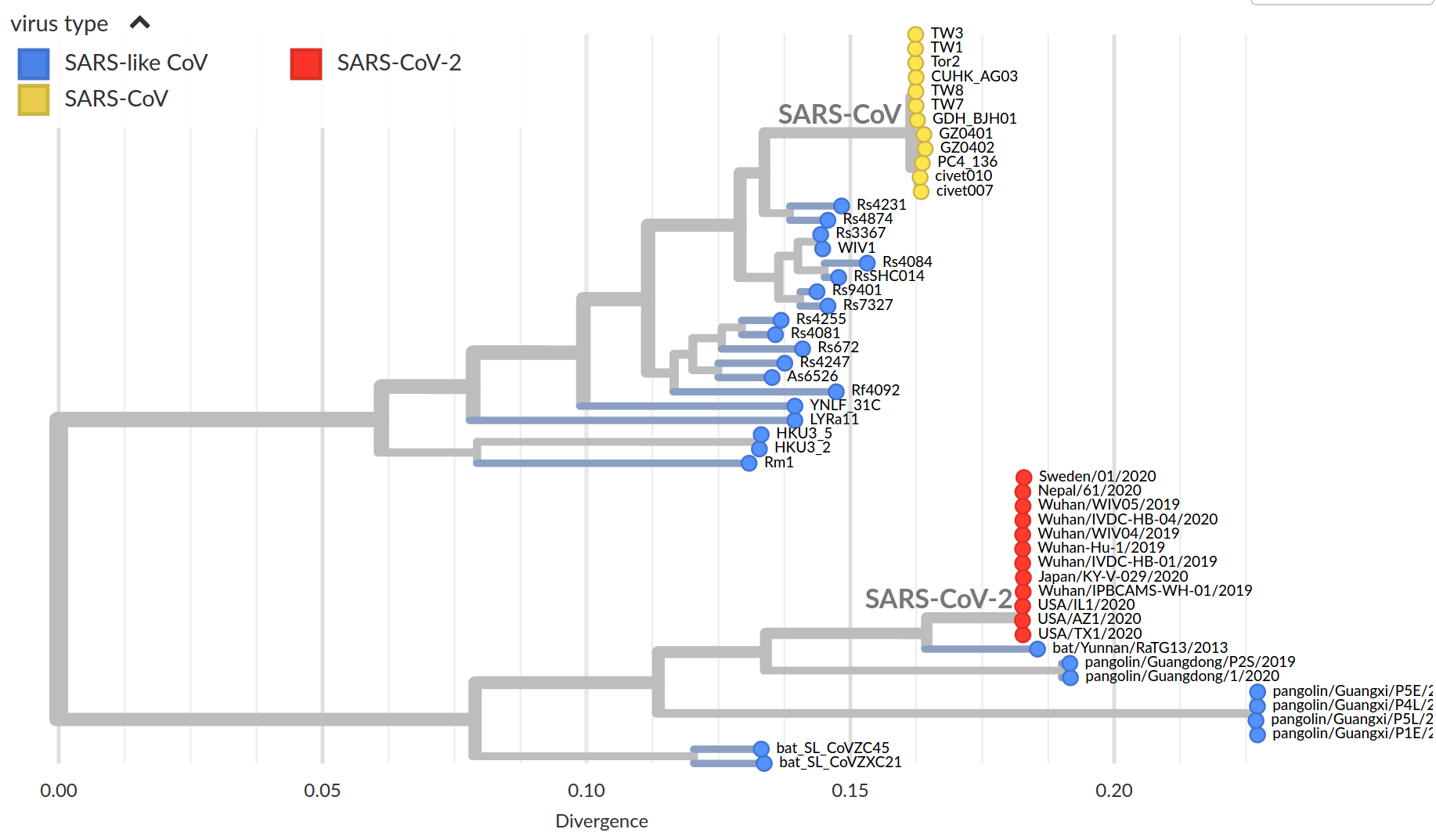 by Trevor Bedford
by Trevor Bedford
Available data on Jan 26 2020
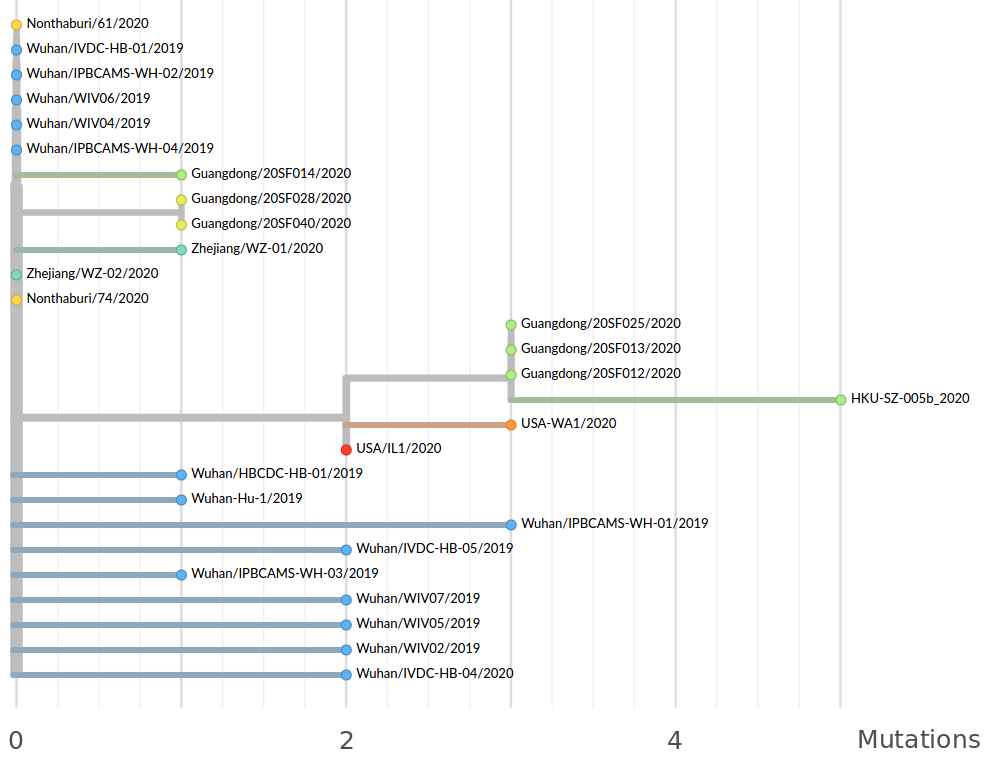
Early genomes differed by only a few mutations, suggesting very recent emergence
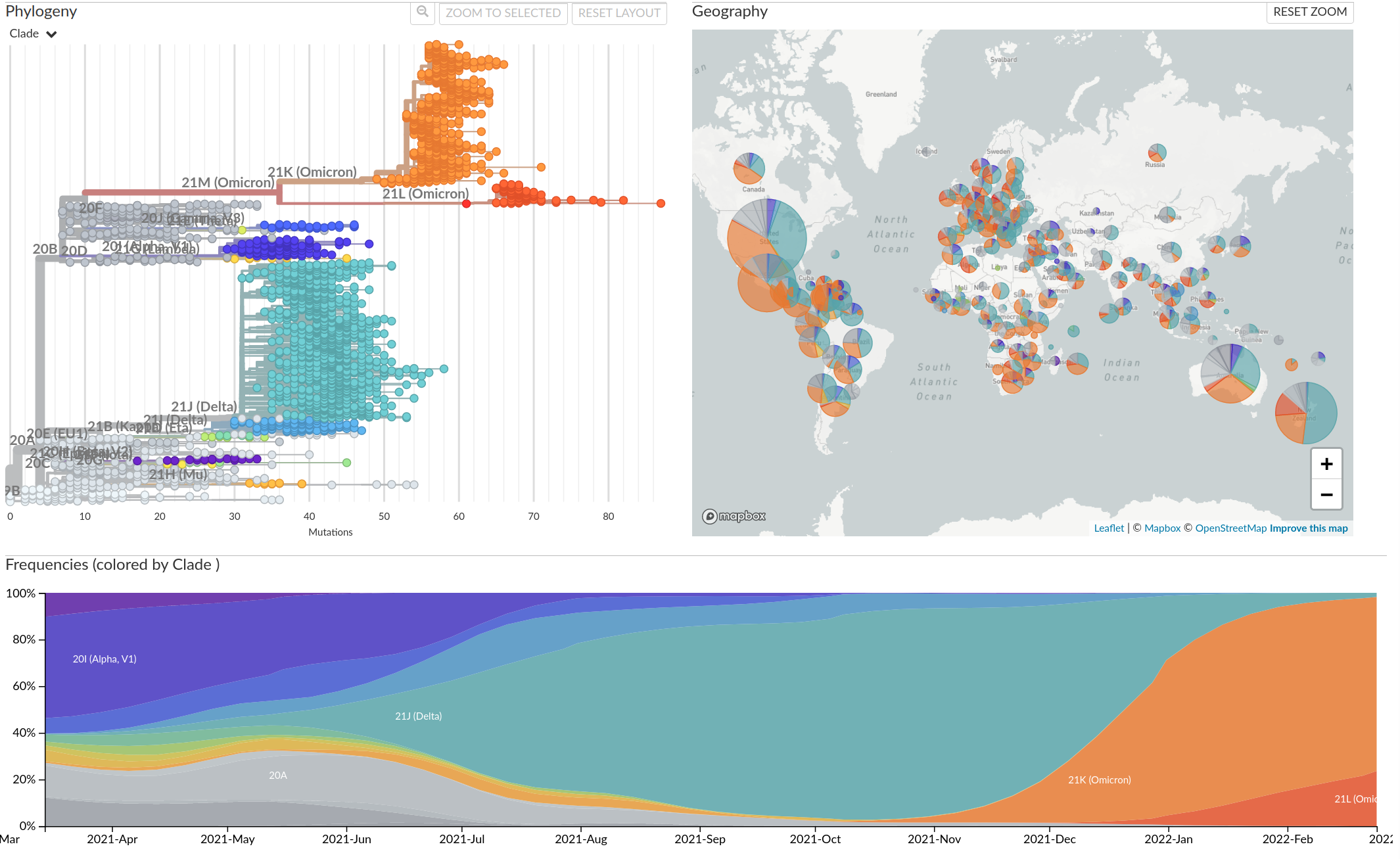
Tracking diversity and spread of SARS-CoV-2 in Nextstrain
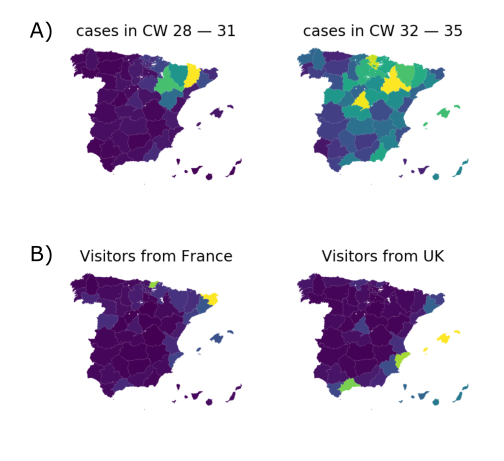
A variant seemed to spread systematically in Summer 2020
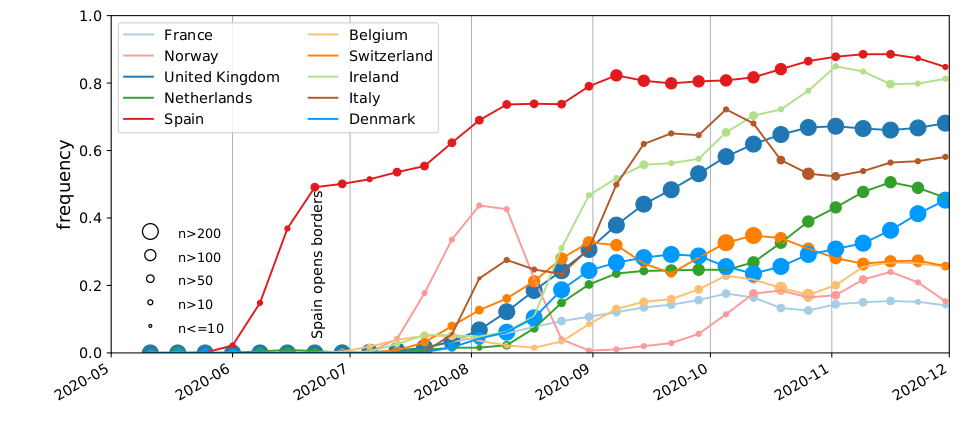
Diversity patterns suggested a large role of holiday travel
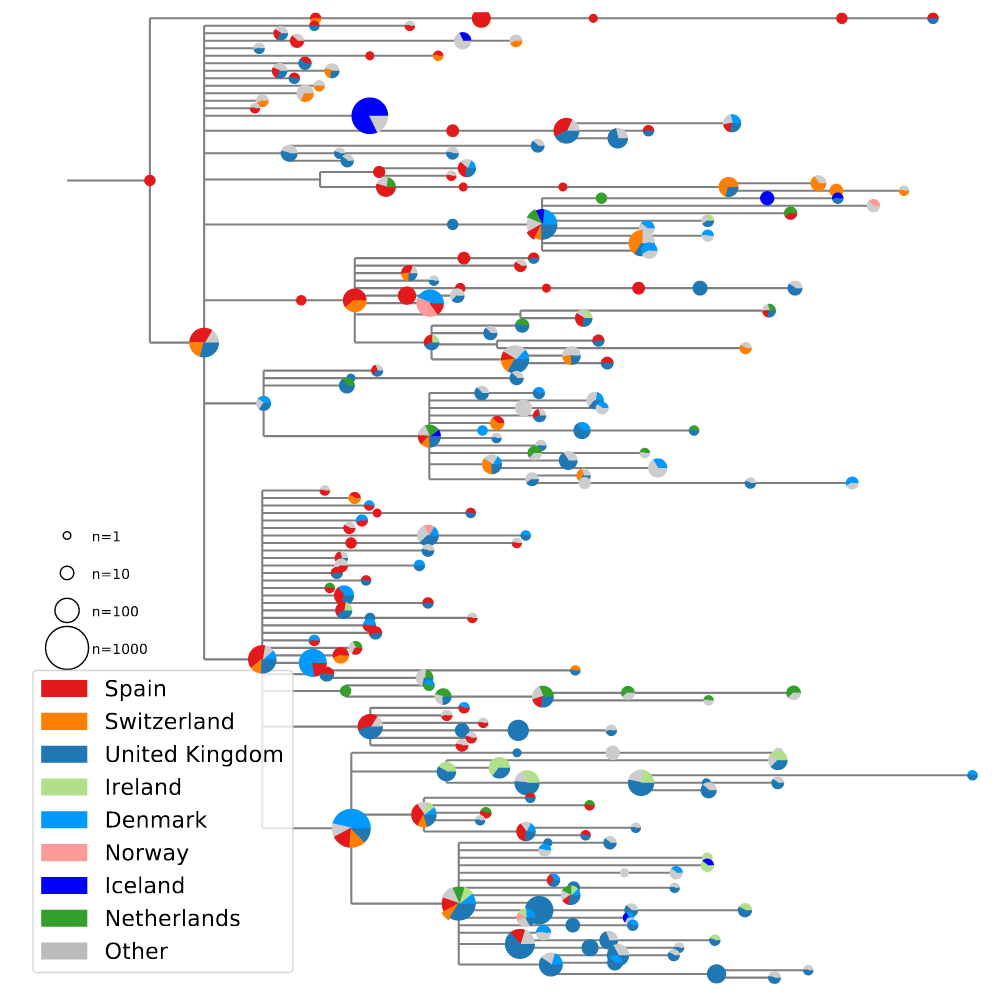
- Spanish diversity mirrored in many European countries
- Suggested many introduction with clusters of similar size
→ travel - A transmission advantage would result in a few dominating introductions
- Screening and quarantine system rather leaky
High case numbers in Spain and high travel volume spread the variant
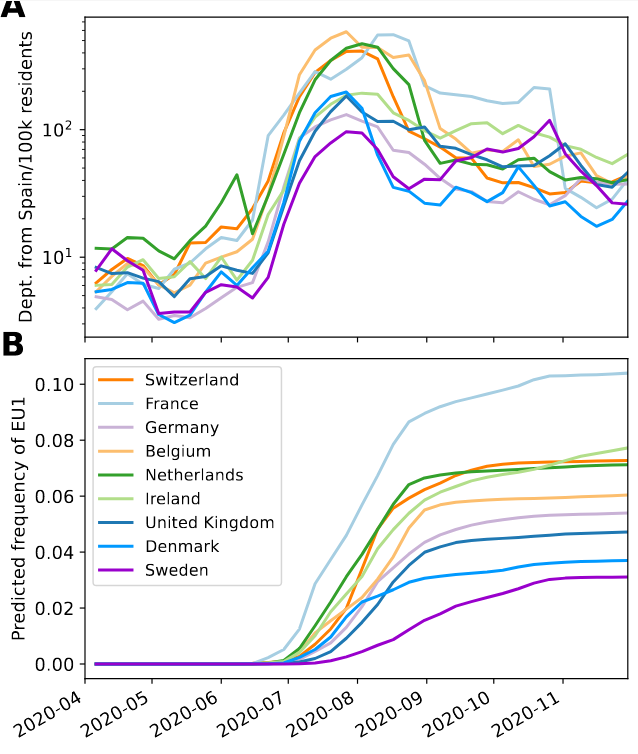
EU1 did not have a strong advantage -- unlike later Variants of Concern
- High incidence differential and high travel volume can drive a variant to dominance
- Travel associated activities and behavior further increases impact
- Onward spread in traveling demographics can be higher
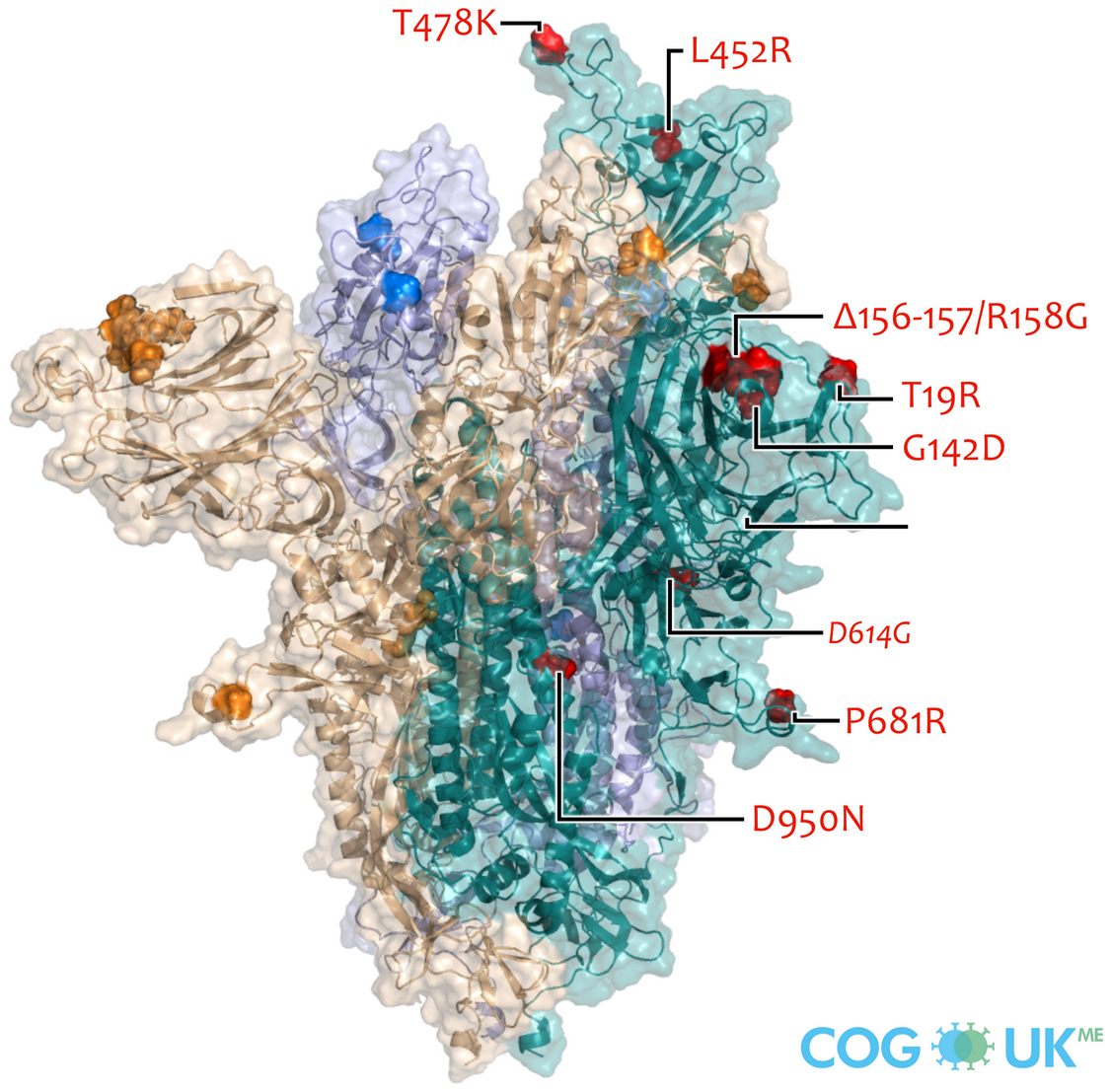
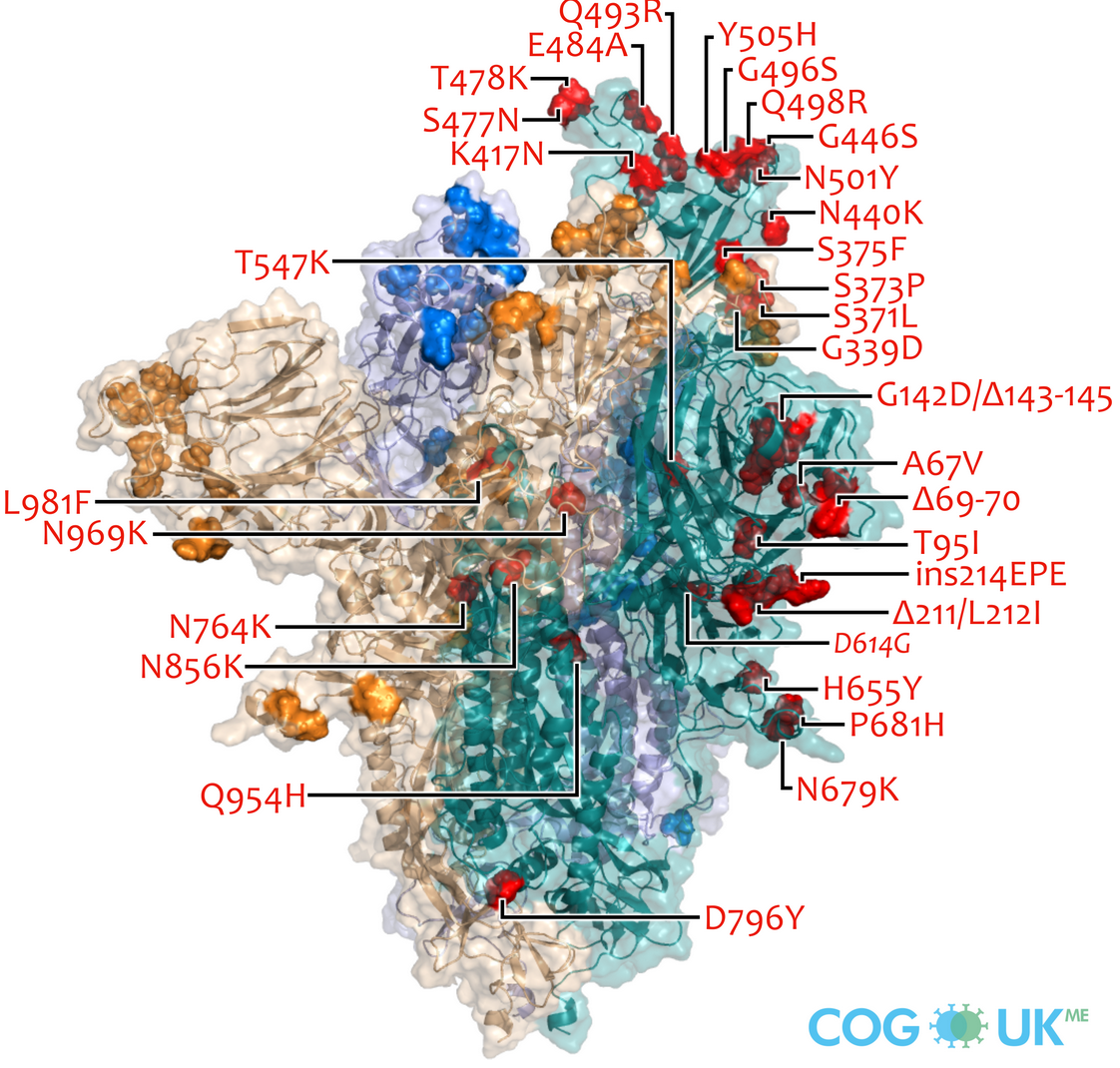
Variants of Concern are characterized by many mutations in S1
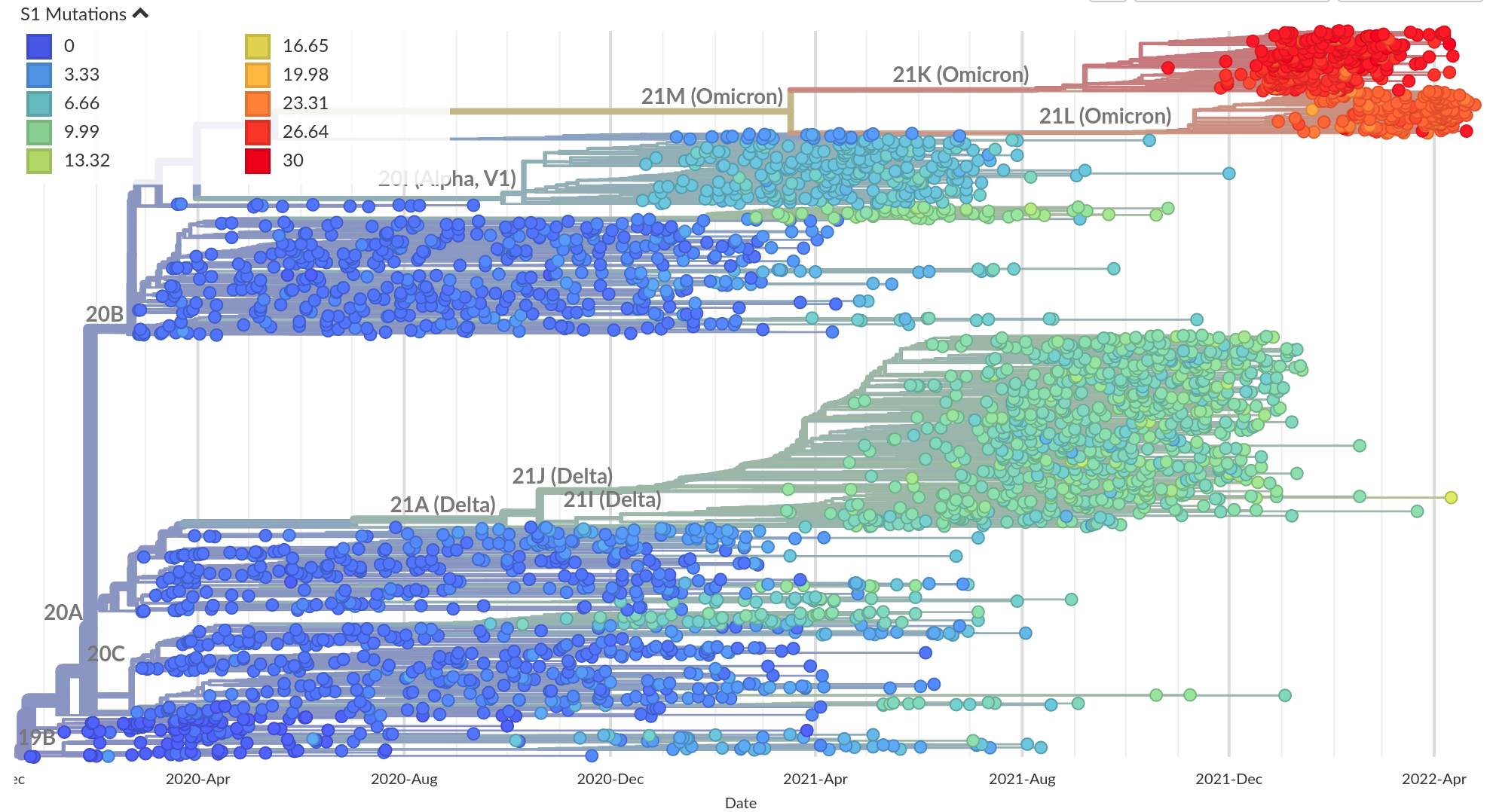
Successful variants are characterized by many mutations in S1
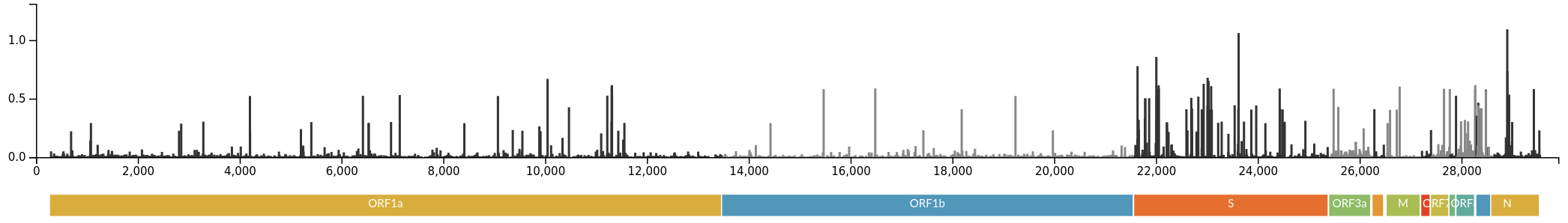
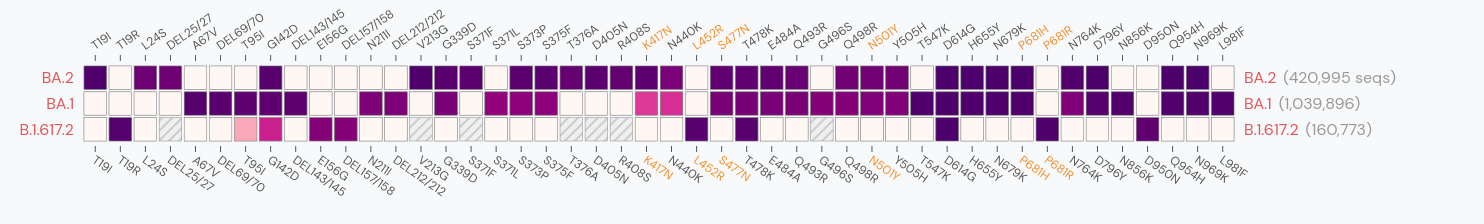
Delta vs Omicron
So far, independent variants have dominated sequentially
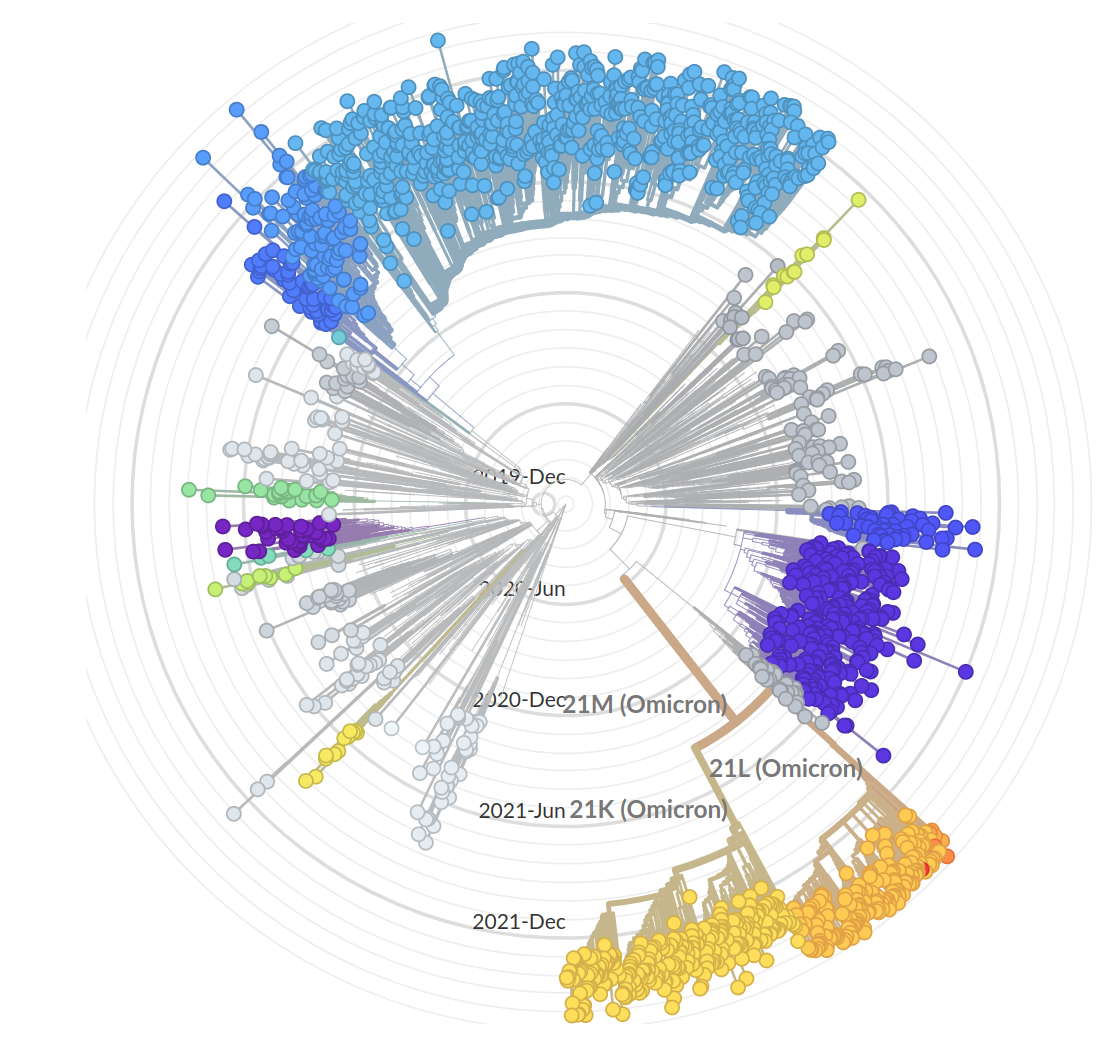
Current Omicron subvariants of interest
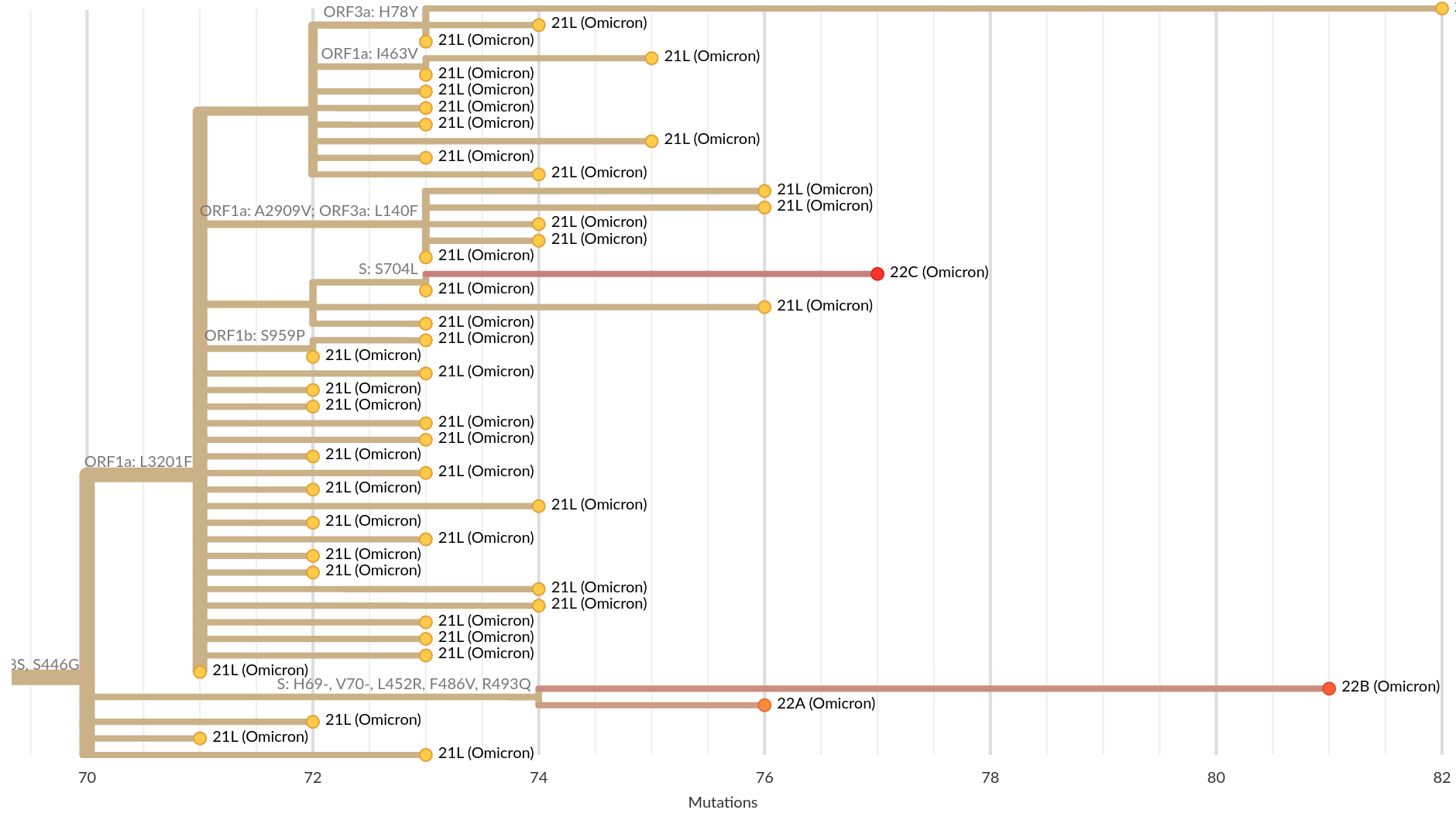
- BA.2 (21L) has essentially taken over.
- BA.4 (22A) and BA.5 (22B) emerged in Southern Africa. Mutations at positions 452 and 486 lead to immune evasion
- BA.2.12.1 (22C) has a mutation at position 452 and is common in the US
Medium term dynamics of SARS-CoV-2 is very uncertain
- Will we start seeing second and third generation variants, as opposed to sister variants?
- Will we the saltatory dynamics with heavily diverged variants continue?
- Will a more diverse immunity landscape slow down future variant dynamics?
- Will waning/antigenic evolution slow down and give rise to annual or even rarer waves?
Going forward, how diverse is SARS-CoV-2 going to be?
Influenza and Theory acknowledgments





- Boris Shraiman
- Colin Russell
- Trevor Bedford
- Pierre Barrat
- Oskar Hallatschek
- All the NICs and WHO CCs that provide influenza sequence data
- The WHO CCs in London and Atlanta for providing titer data


SARS-CoV-2 acknowledgements


- Emma Hodcroft (now in Bern)
- Moira Zuber (Basel)
- Iñaki Comas and Fernando Gonzalez-Candelas, Valencia
- Martina Reichmuth and Christian Althaus (Bern)
- Tanja Stadler, Sarah Nadeau, Tim Vaughan at ETH
- Alberto Hernando and David Matteo at Kido Dynamics
- Jesse Bloom, Katherine Crawford at Fred Hutch
- David Veesler, Alex Walls, Davide Corti, John Bowen at UW

Acknowledgments
Trevor Bedford and his lab -- terrific collaboration since 2014

especially James Hadfield, Emma Hodcroft, Ivan Aksamentov, Cornelius Roemer, Moira Zuber, and John Huddleston
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID

