Transmission, escape, and resistance: how human pathogens adapt and change
Richard Neher
Biozentrum & SIB, University of Basel
slides at neherlab.org/202207_Berlin.html

What sustains endemic circulation? Can we predict/understand?
Continuous circulation requires a continuous "supply" of susceptibles
- Birth of immunologically naive individuals
- Waning of pre-existing immunity
- Antigenic change of the virus
- Non-sterilizing immunity → persistence via asymptomatic acute infections
Measles Virus (pre vaccine)
- Extremely rapid transmission among naive children
- Long-lived immunity → rare re-infection
Influenza Virus (and many others!)
- Frequent reinfection of adults
- Combination of waning and antigenic evolution
- Approximate equilibrium between immunity and infections
Positive tests for influenza in the USA by week
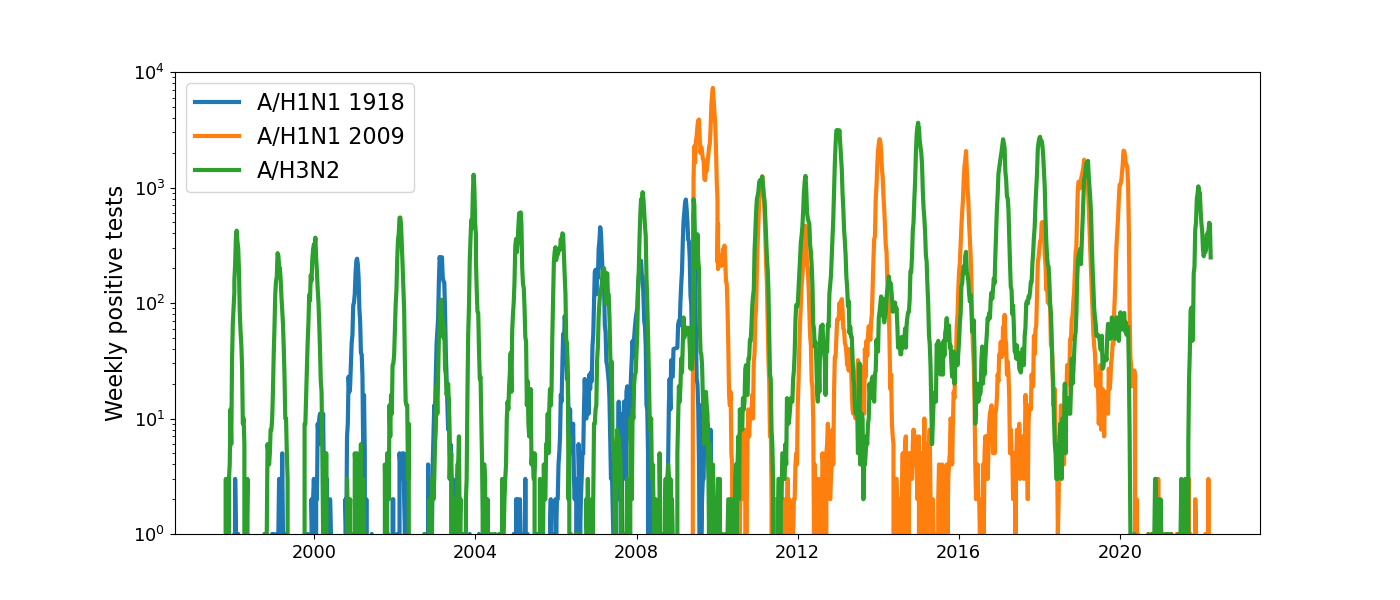 Data by the US CDC
Data by the US CDC
Human seasonal influenza viruses

Influenza A/H3N2
Genomic analysis to reconstruct pathogen spread and evolution
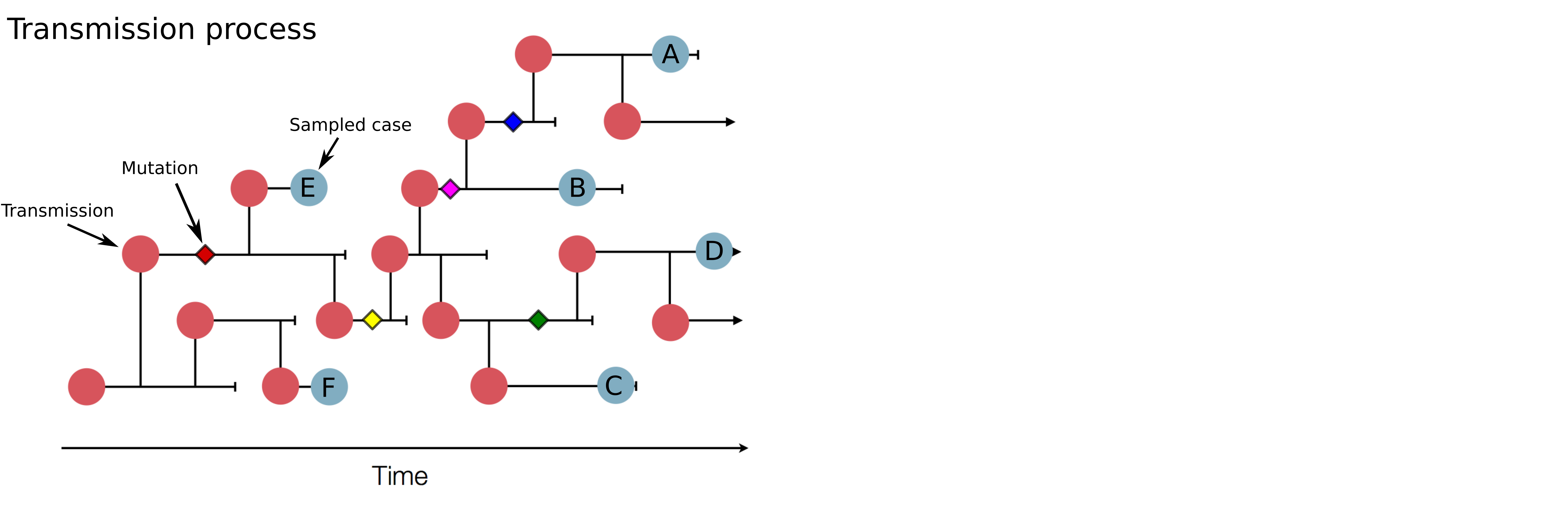
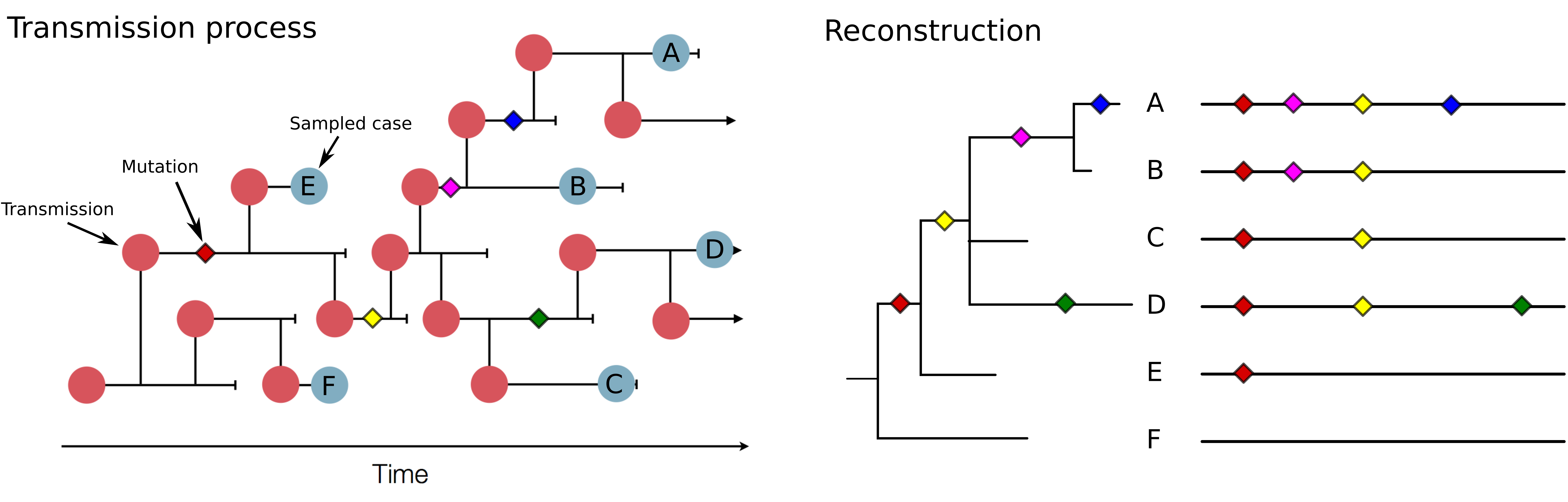
Track how pathogens spread: clusters, introductions, etc
Link genotypic and phenotypic changes: immune escape, drug resistance, host adaptation



- Influenza viruses evolve to avoid human immunity
- Vaccines need frequent updates

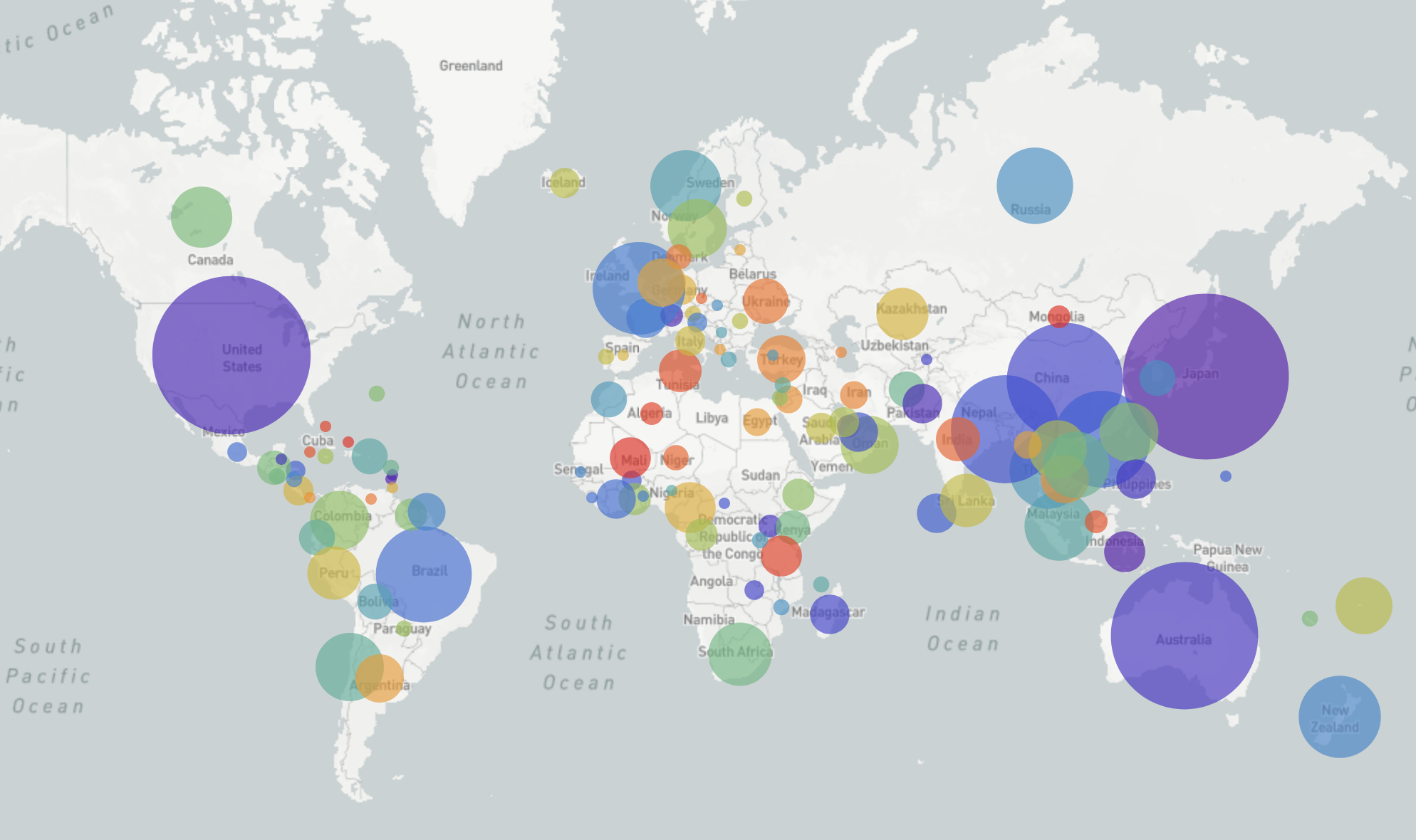
Nextstrain.org: real-time analyses of influenza virus evolution
RN et al, 2014; RN & Bedford, 2015; Huddleston et al, 2020
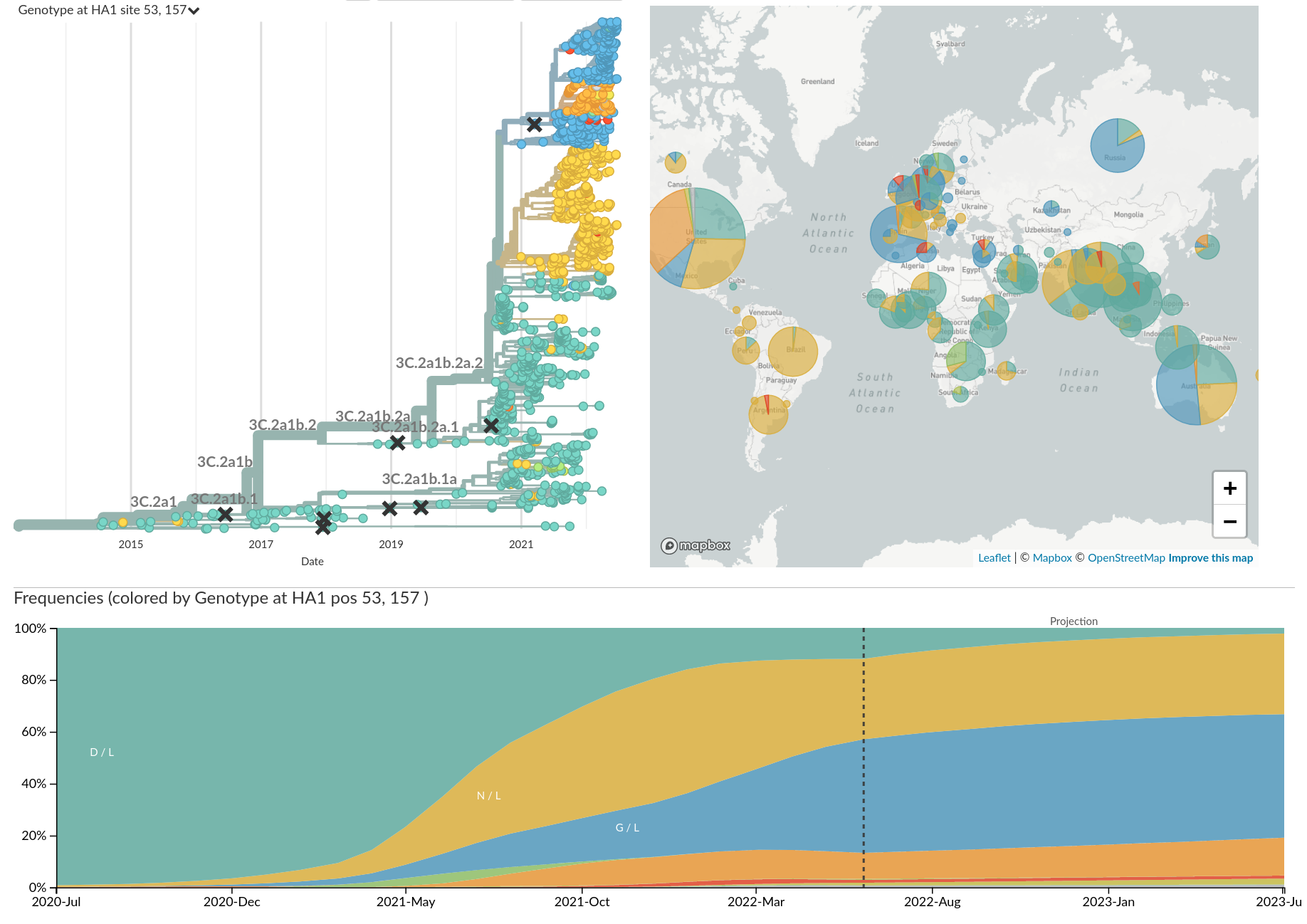
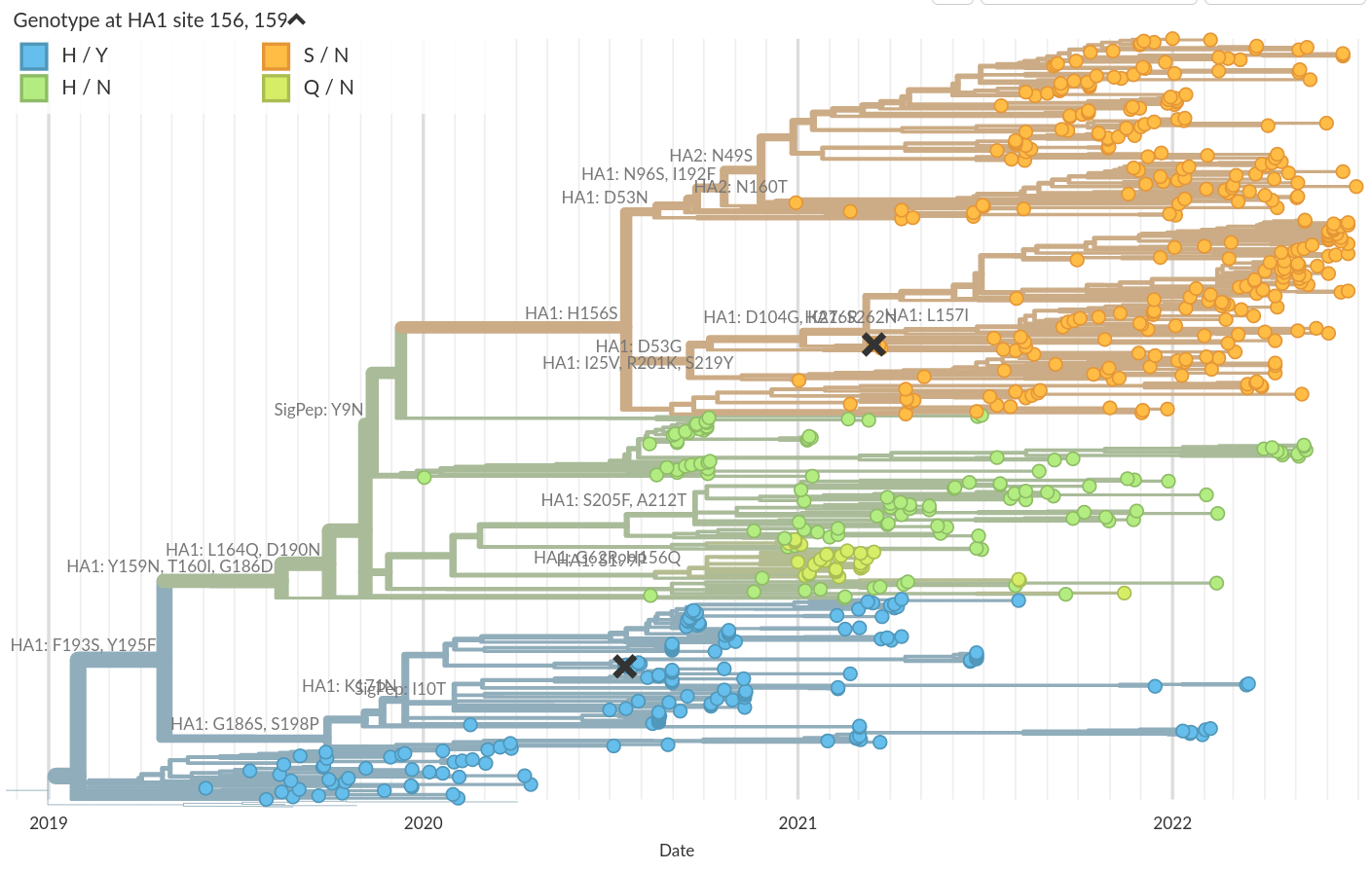
Genotypic changes
Antigenic changes via hemagglutination inhibition assays
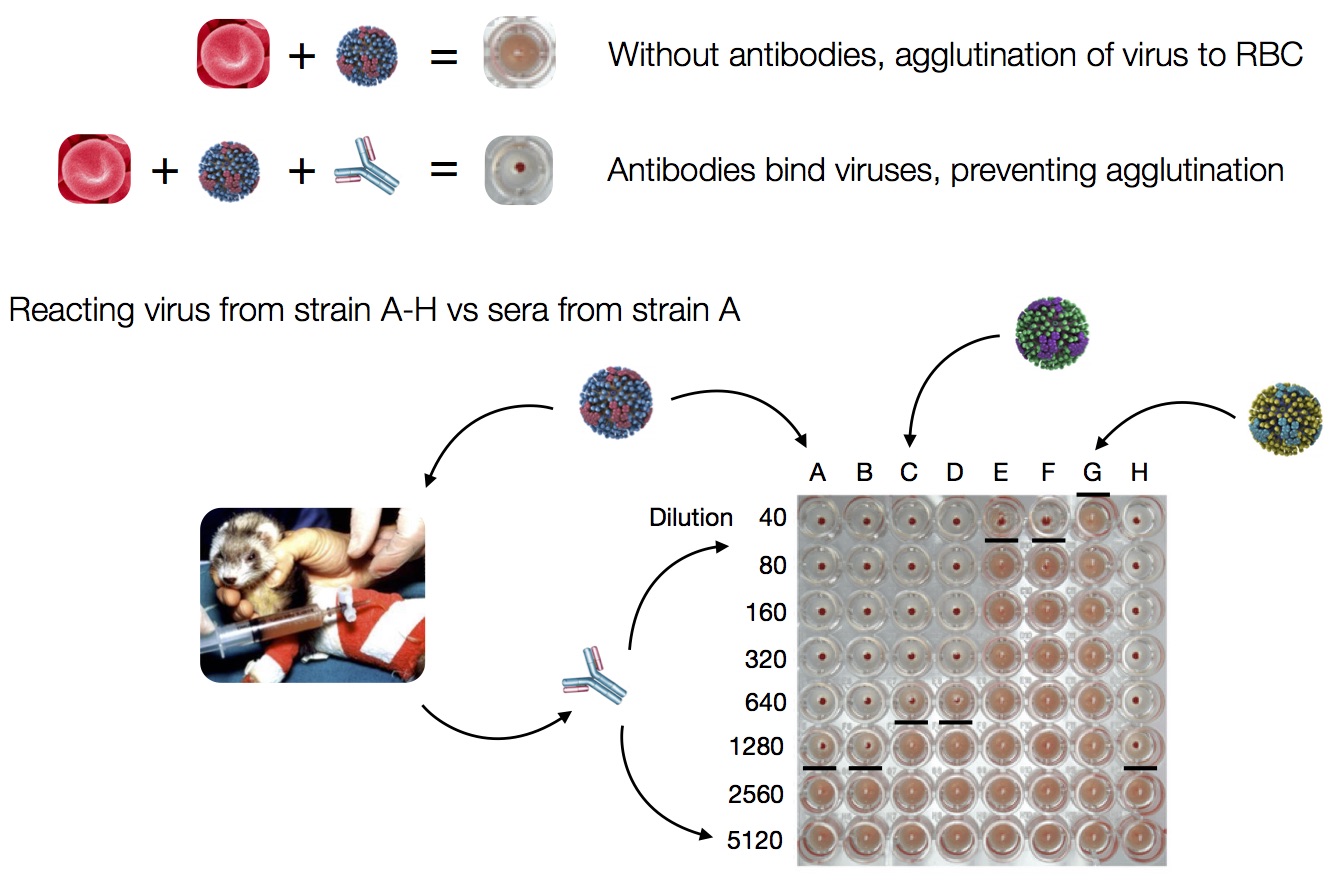
Antigenic distance tables
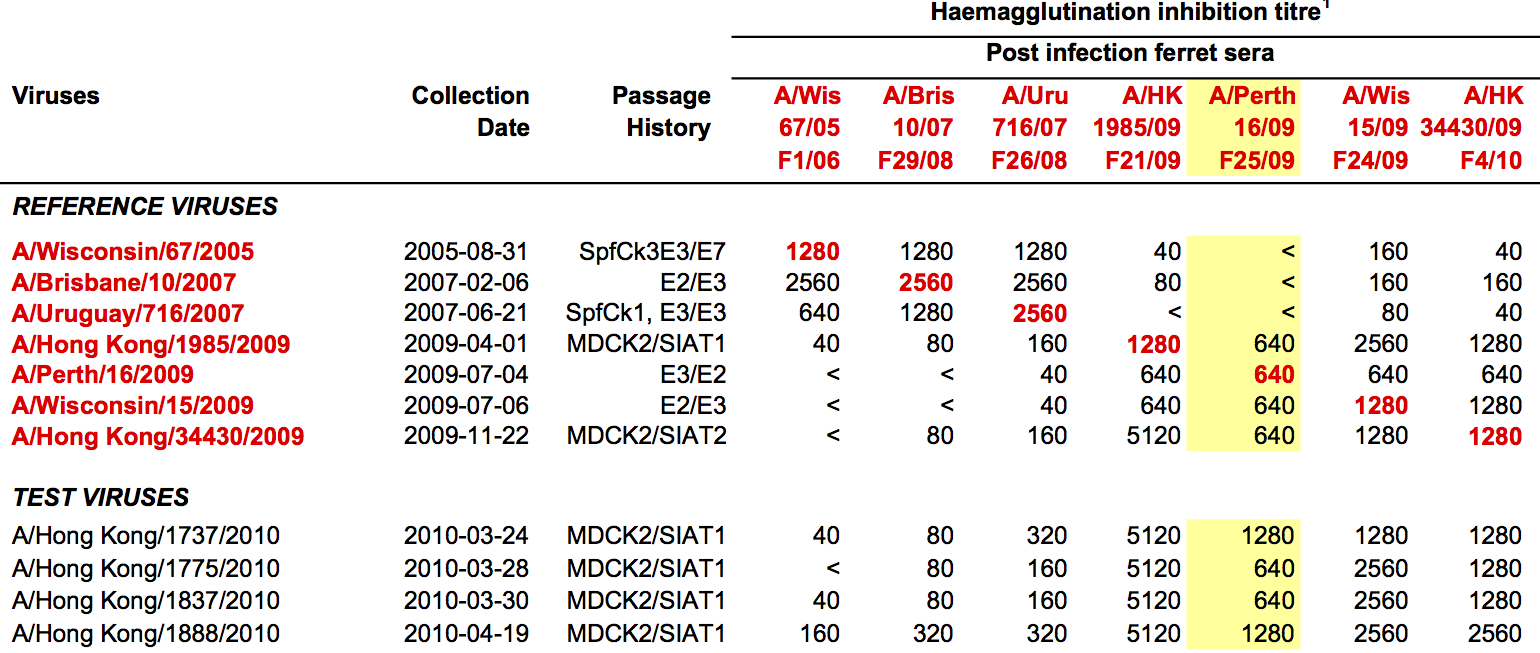
- Long list of distances between sera and viruses
- One-to-one link between sera and the virus used to infect the ferret
- Tables are sparse, only closely related pairs covered.
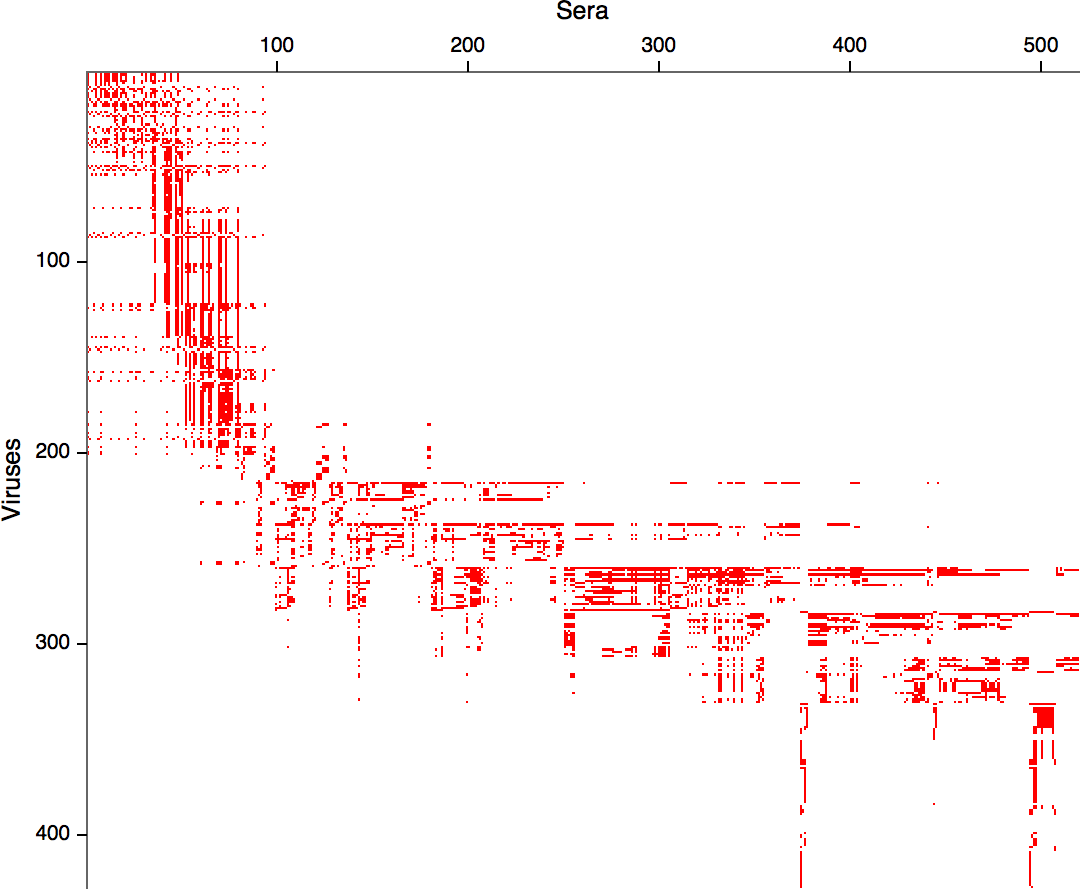
Integrating antigenic data (ferret serology) and molecular evolution
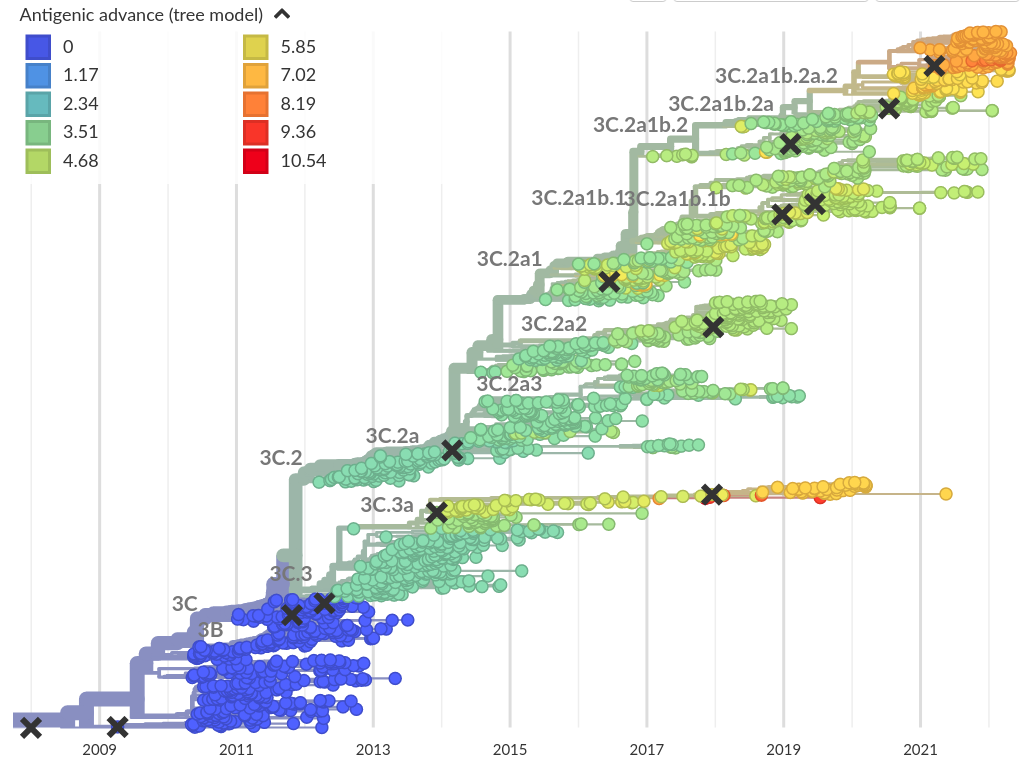
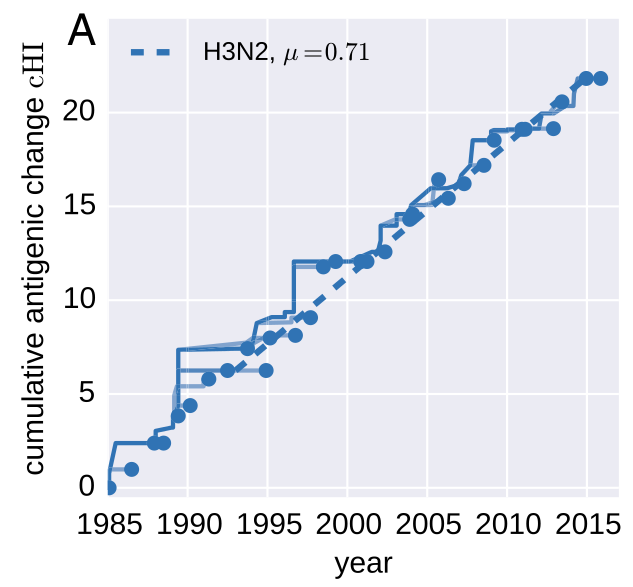
- ~2-fold reduction in titers per year (Omicron vs Ancestral: ~32-fold)
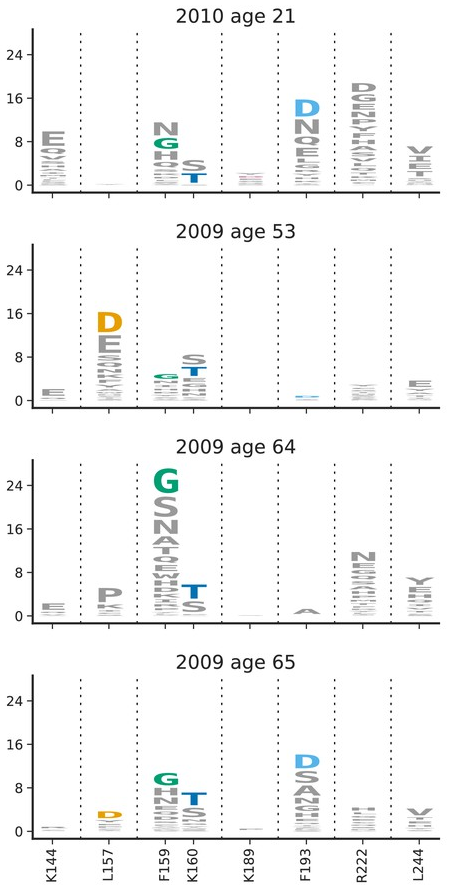
- Link antigenic changes with specific mutations mostly near the receptor binding site
- Extrapolate antigenic properties to un-characterized viruses
Prediction of successful variants to optimize vaccine composition

RN et al, 2014; RN & Bedford, 2015; Huddleston et al, 2020; Barrat-Charlaix et al, 2020
A/H3N2 variants replace each other every ~3 years
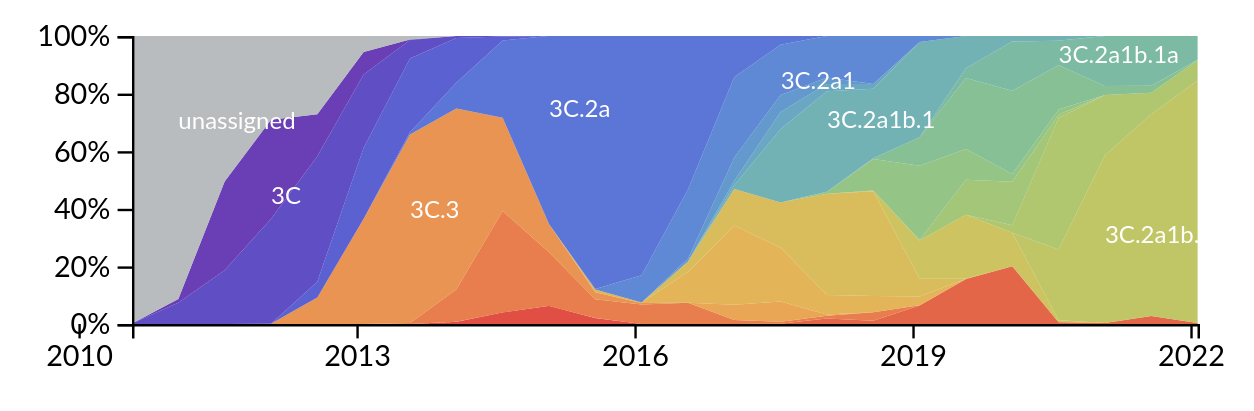
SARS-CoV-2
Tracking diversity and spread of SARS-CoV-2 in Nextstrain
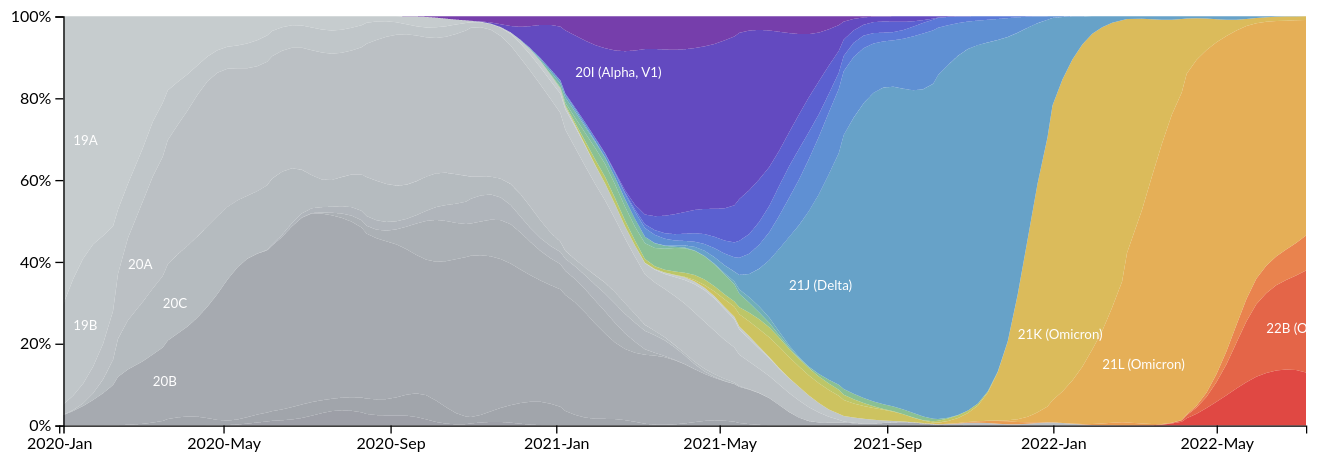
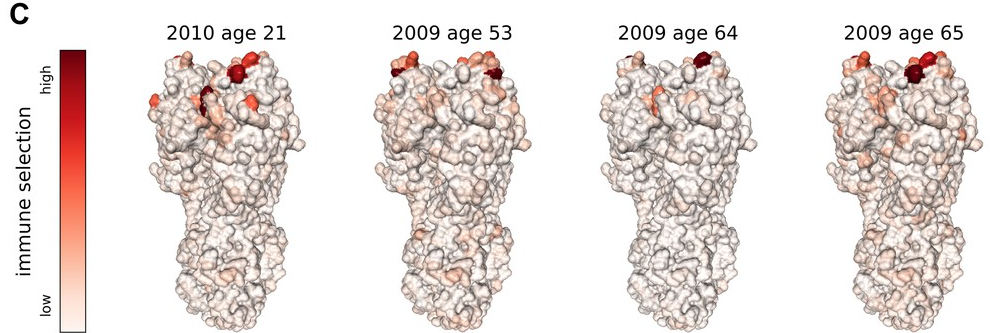
Successful variants are characterized by many mutations in S1
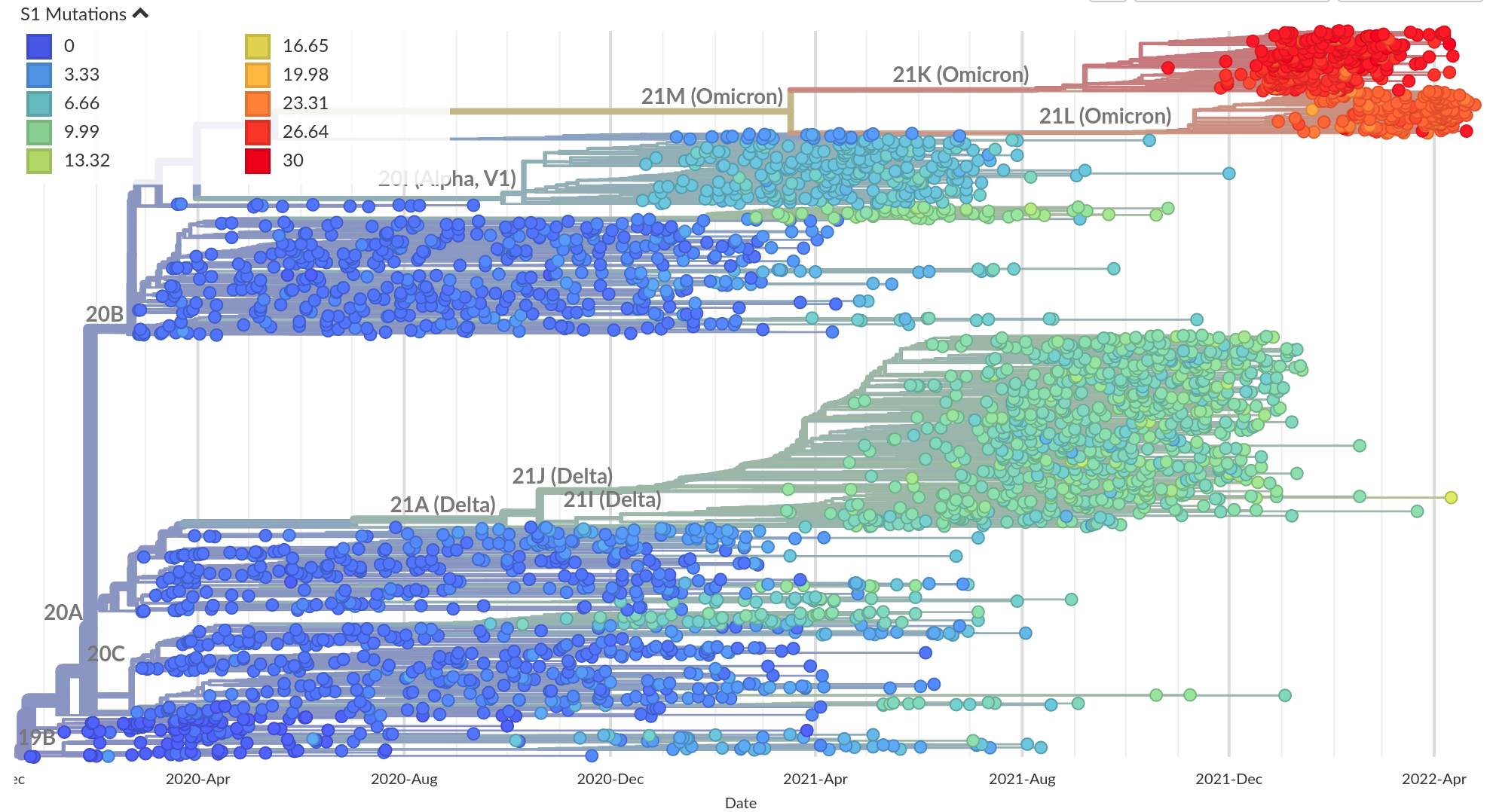
Remarkable patterns of rapid adaptation
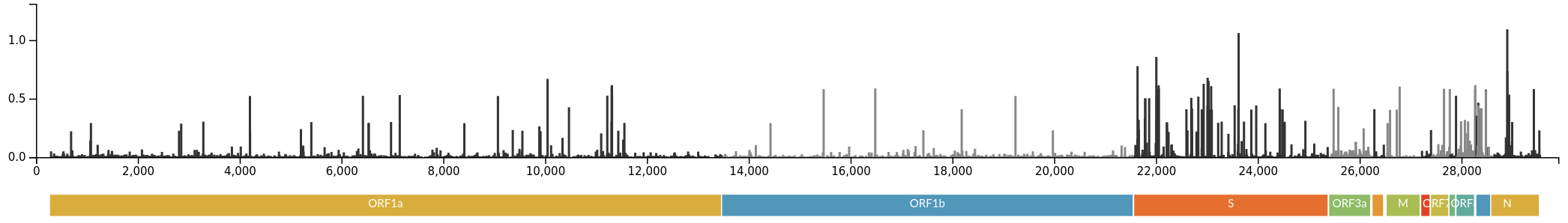
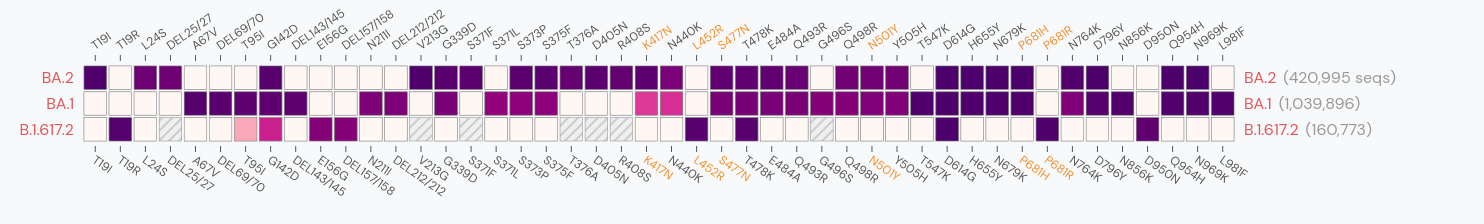
Gradual shift from selection on transmission to immune escape
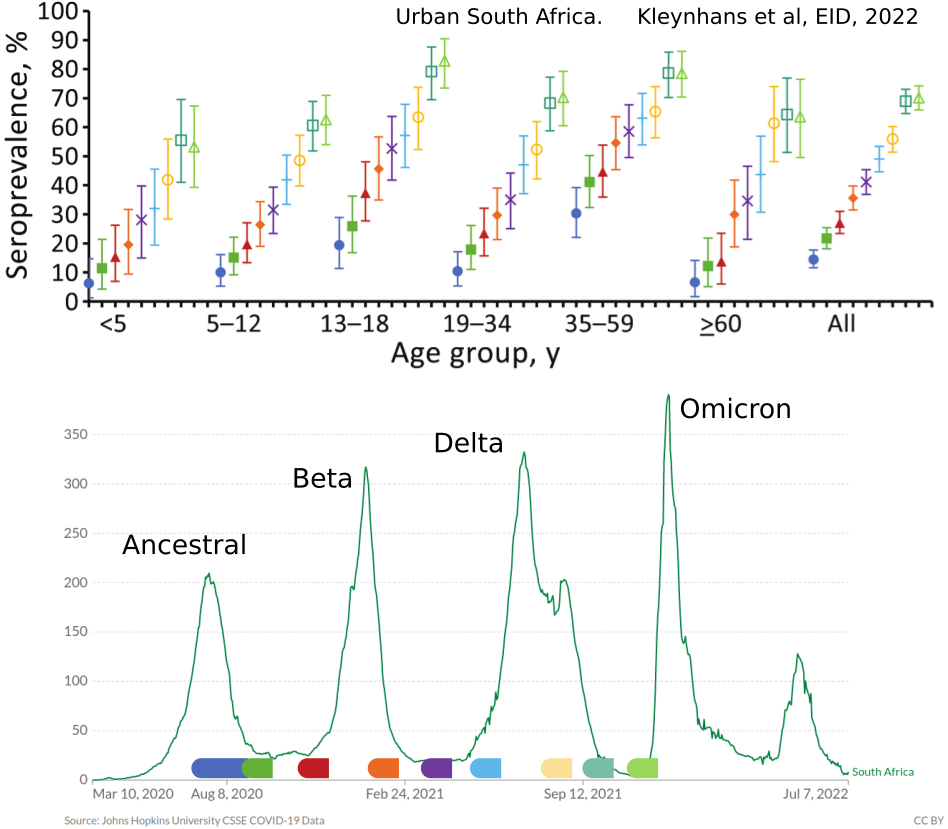
- Until early-2021, seroprevalence was low to moderate
- Delta infections and vaccination resulted in high seroprevalence in 2021
- Variant success now is dominated by immune escape
So far, independent variants have dominated sequentially
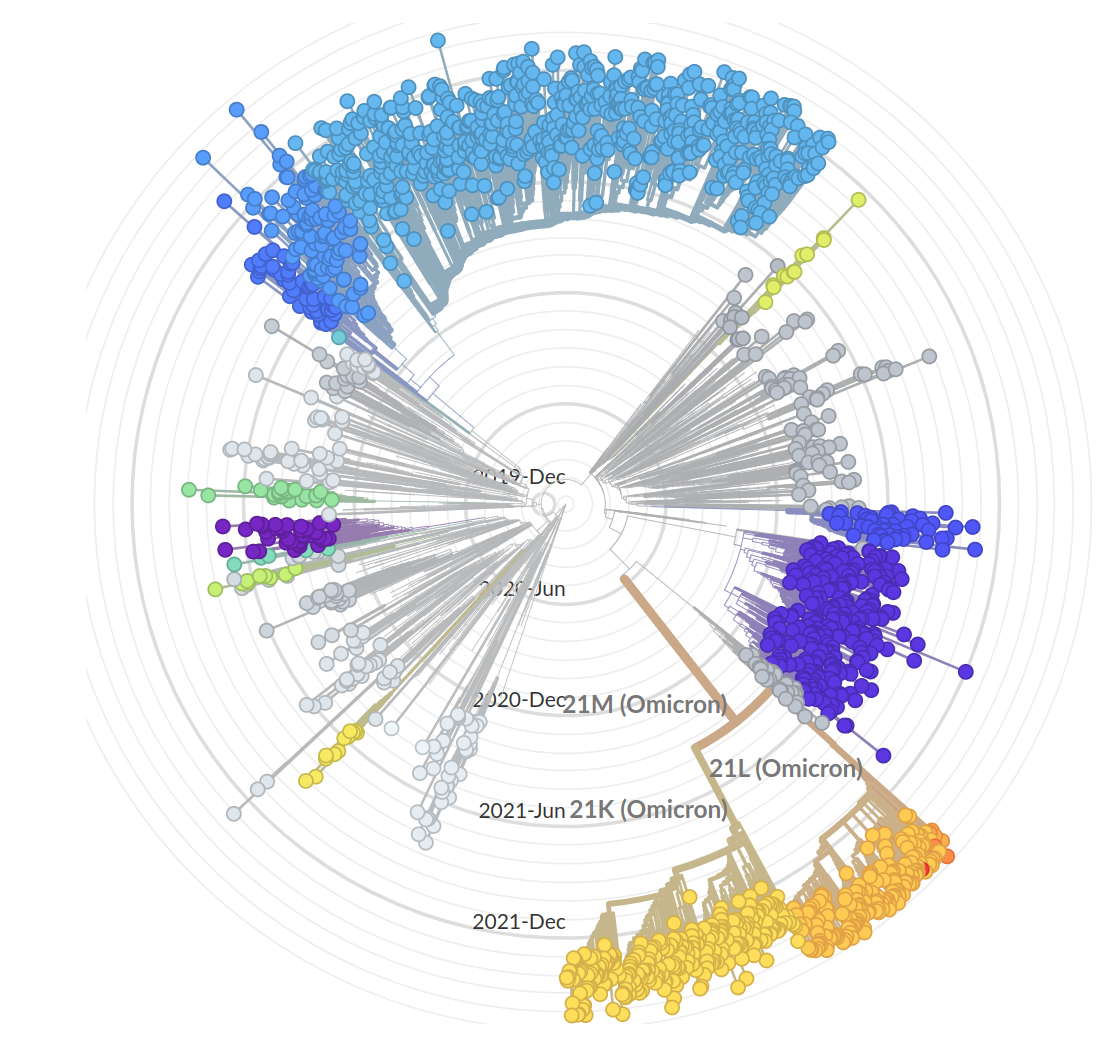
- Variant emergence likely through chronic infections
- Strong dichotomy (until 2022): dramatic changes between variants, slow and steady within variants
- Omicron variants are more dynamic
- With BA.4/5 and BA.2 subvariants, we start to see second generation variants
What is driving this?
Can we predict?
To predict, we need to quantify selection by immunity
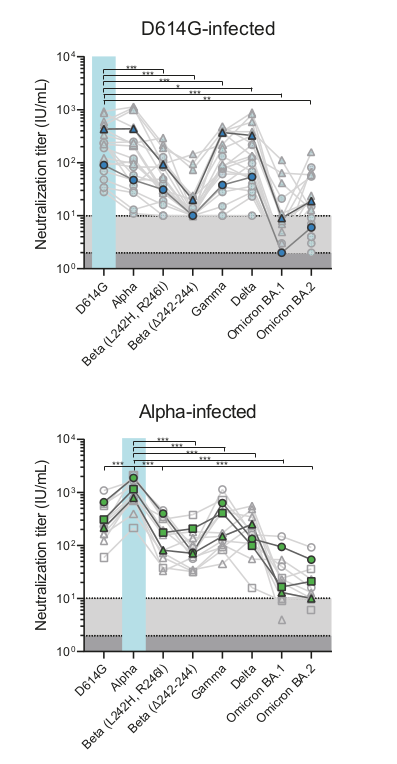
- Given a population immunity "landscape", how much escape is caused by which mutation?
- How variable are individual immune responses?
- How does exposure history shape neutralization of different variants?
- What is the contribution of chronic vs acute infections?
- Does escape during chronic infection mediate inter-individual escape? It did for Omicron, but will this stay that way?
- What is the contribution of chronic infections in other viruses?
Host-microbe co-evolution beyond viruses
RNA viruses: mutations and homologous recombination


Bacteria: pick and choose from large pangenomes
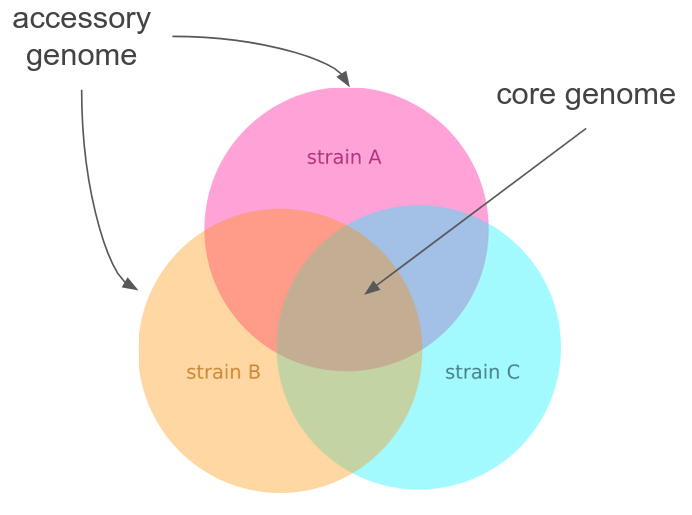
- Complex patterns of horizontal transfer of homologous and non-homologous sequence
- Host-pathogen interactions and resistance controlled by the accessory genome

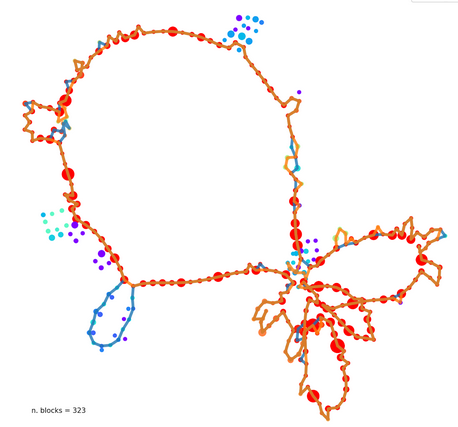
Microbial pangenome graphs
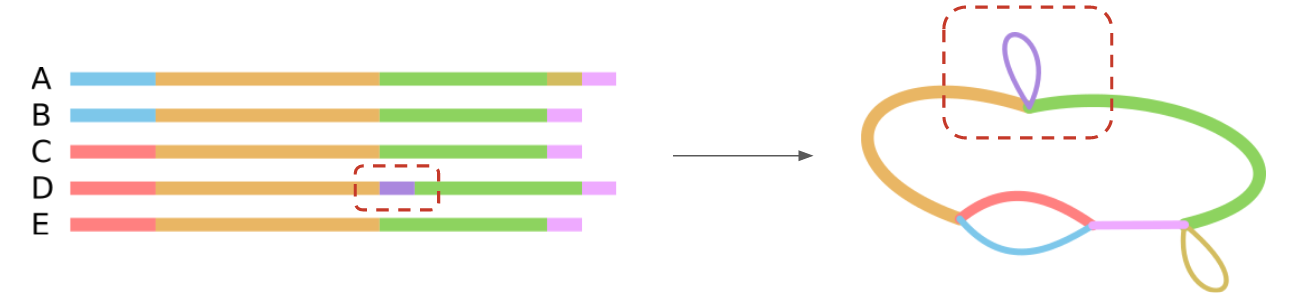
- Efficient method to construct pan-genome graphs of dozens to hundreds of (similar) genomes
- Capture genome similarity, content, and structure
- Breakpoints in the graph correspond to evolutionary events
Microbial pangenome graphs
Klebsiella pneumonia genomes
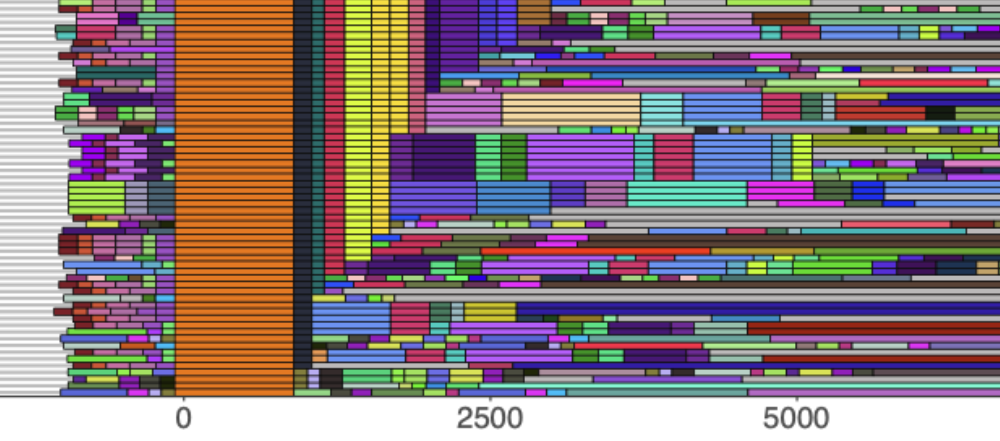
New Delhi beta-lactamase carrying plasmids
Summary and outlook
- RNA viruses:
- diversity and evolution well covered
- rapid phenotyping of relevant traits challenging
- Immunological diversity of humans:
- many basic open questions how repeated infections shapes the repertoire
- important implications for prediction of evolution to improve vaccines
- what we are doing for SARS-CoV-2 and influenza is inadequate
- we can measure 100s of antigens against 100s of sera in longitudinal cohorts
- Bacteria: challenges in diversity and evolution are on the pathogen side
- vast pan-genomes of largely unknown function
- extensive horizontal transfer
- long-read sequencing can resolve genomes now at scale
Acknowledgments
Trevor Bedford and his lab -- terrific collaboration since 2014

especially James Hadfield, Emma Hodcroft, Ivan Aksamentov, Cornelius Roemer, Moira Zuber, and John Huddleston
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID


Acknowledgments



- Nicholas Noll
- Marco Molari
- Liam Shaw
