Evolution and spread of Influenza, HIV, and SARS-CoV-2
Richard Neher
Biozentrum, University of Basel
slides at neherlab.org/202212_CBB.html
Human seasonal influenza viruses

Positive tests for influenza in the USA by week
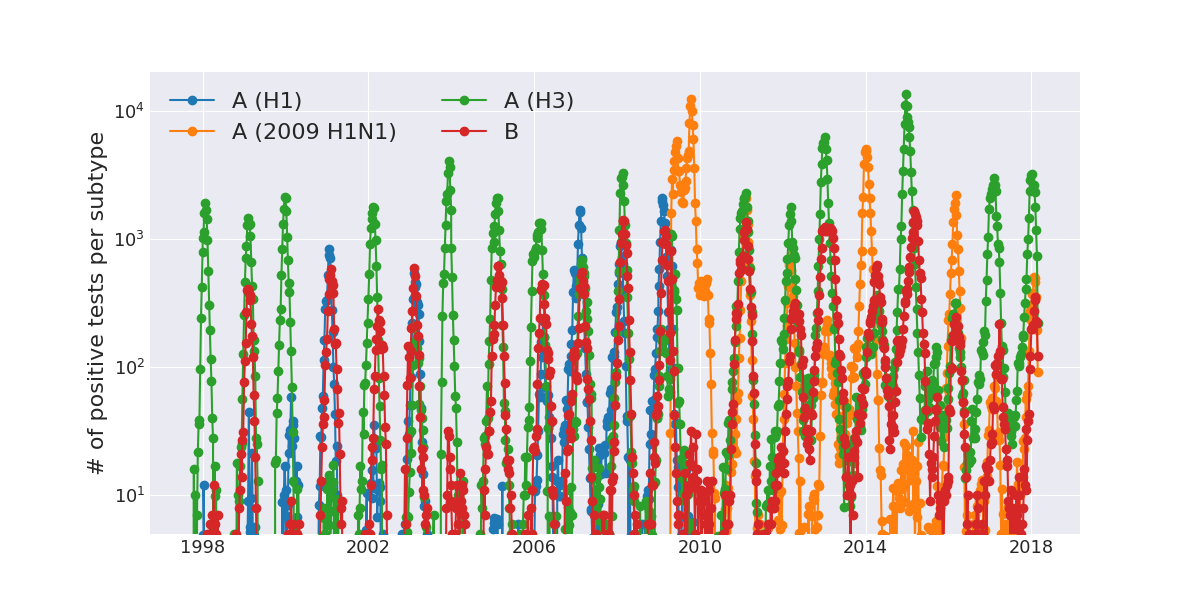 Data by the US CDC
Data by the US CDC
Virus genomes change rapidly through time
A/Brisbane/100/2014
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
A/Brisbane/1000/2015
GGATAATTCTATTAACCATGAAGACTATTATTGCTTT...
A/Brisbane/1/2017
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
... hundreds of thousands of sequences...
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
A/Brisbane/1000/2015
GGATAATTCTATTAACCATGAAGACTATTATTGCTTT...
A/Brisbane/1/2017
GGATAATTCTATTAACCATGAAGACTATCATTGCTTT...
... hundreds of thousands of sequences...
Genome sequences record the spread of pathogens




in SARS-CoV-2 a new mutation accumulates about every 2 weeks



- Influenza viruses evolve to avoid human immunity
- Vaccines need frequent updates

Predicting evolution

Given the branching pattern:
- can we predict fitness?
- pick the closest relative of the future?
Fitness inference from trees
$$P(\mathbf{x}|T) = \frac{1}{Z(T)} p_0(x_0) \prod_{i=0}^{n_{int}} g(x_{i_1}, t_{i_1}| x_i, t_i)g(x_{i_2}, t_{i_2}| x_i, t_i)$$
RN, Russell, Shraiman, eLife, 2014
Prediction of the dominating H3N2 influenza strain
- no influenza specific input
- how can the model be improved? (see model by Luksza & Laessig)
- what other context might this apply?
Real-time tracking of SARS-CoV-2
- thousands of new sequences every day
- more than >14M sequences right now
- comprehensive analysis require hours to days to complete
→ requires continuous analysis and easy dissemination
→ interpretable and intuitive visualization
nextstrain.org
joint project with Trevor Bedford & his lab
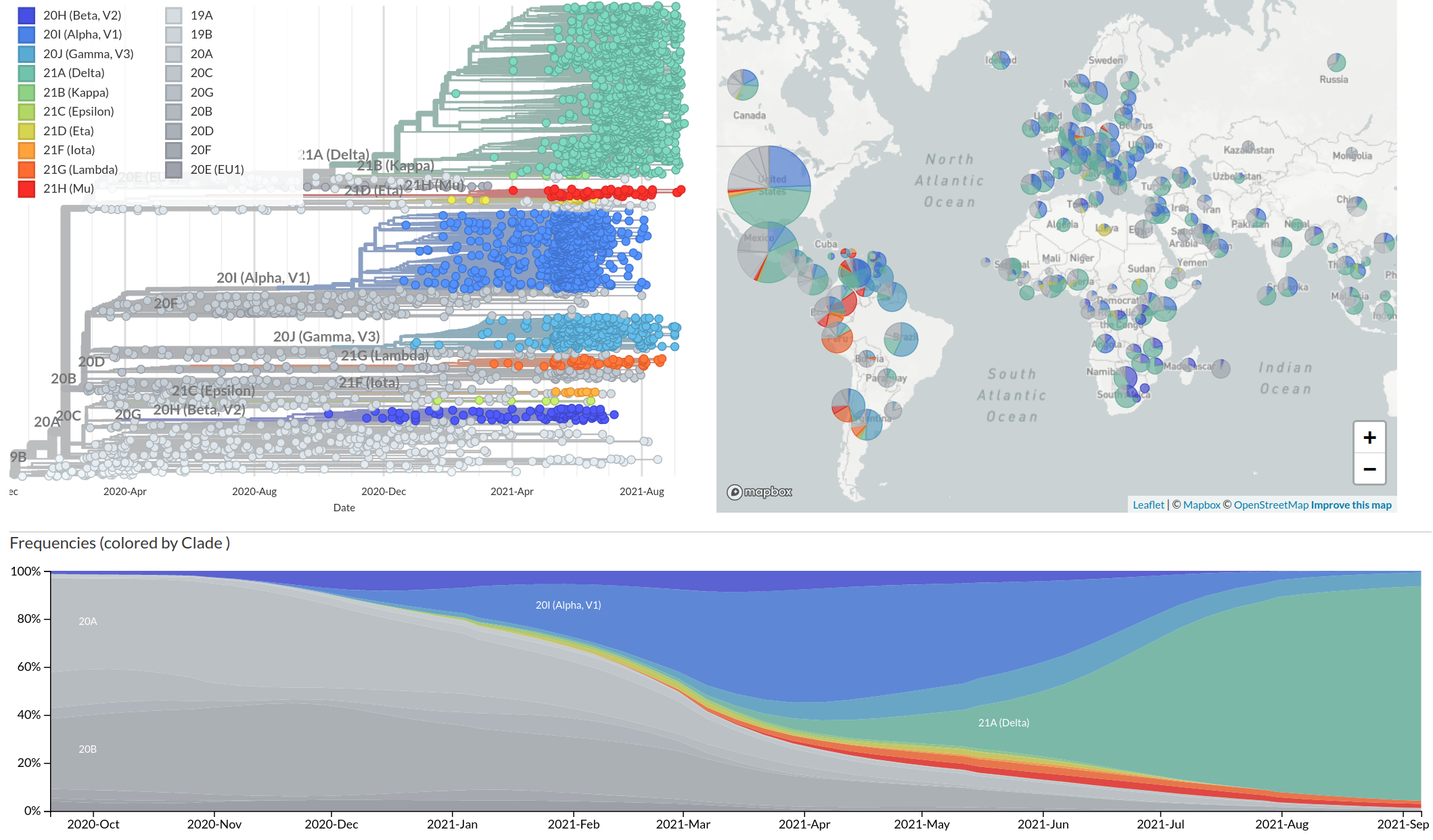
Emergence and dominance of SARS-CoV-2 variants
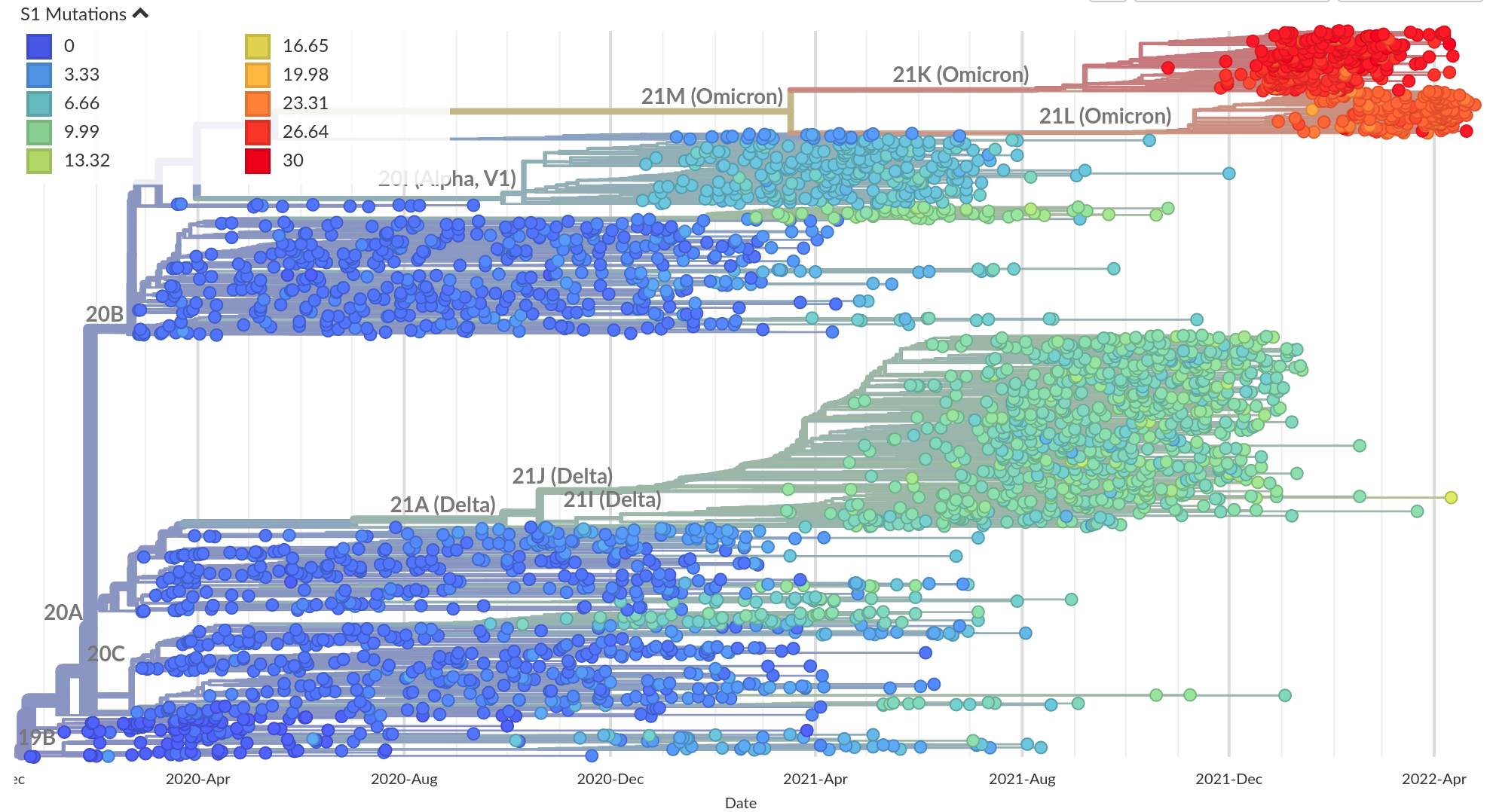
Successful variants are characterized by many mutations in S1
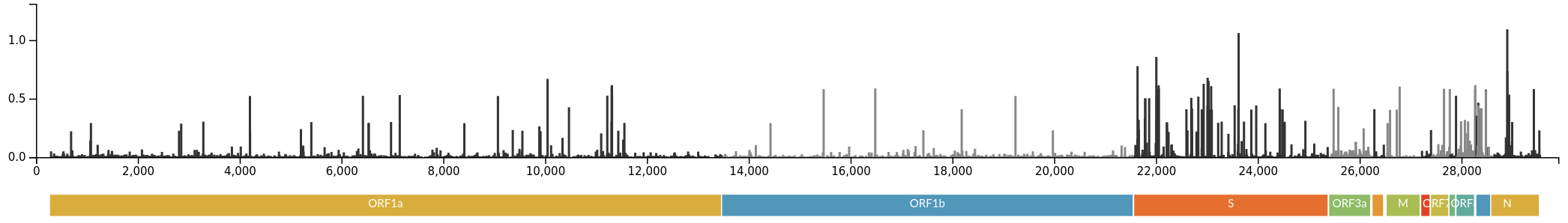
Remarkable patterns of rapid adaptation
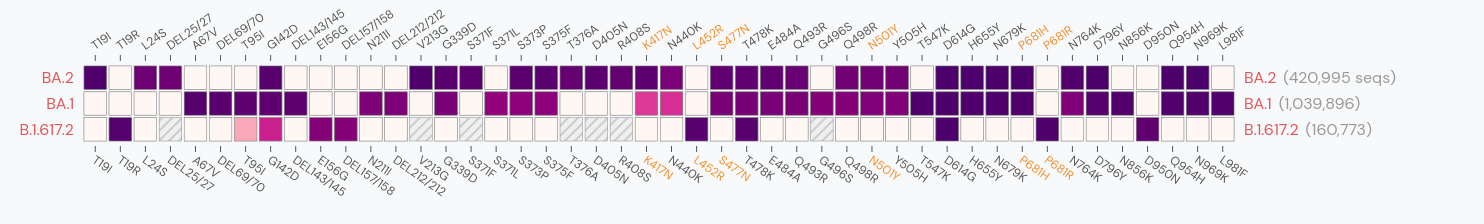
Gradual shift from selection on transmission to immune escape
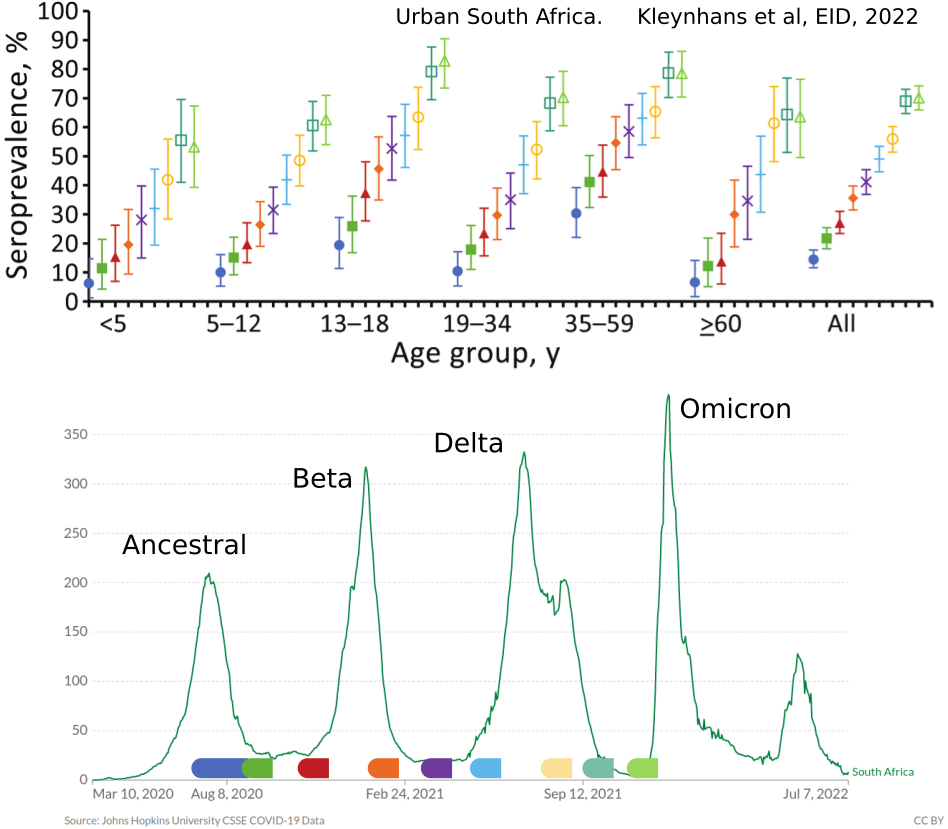
- Until early-2021, seroprevalence was low to moderate
- Delta infections and vaccination resulted in high seroprevalence in 2021
- Variant success now is dominated by immune escape
So far, independent variants have dominated sequentially
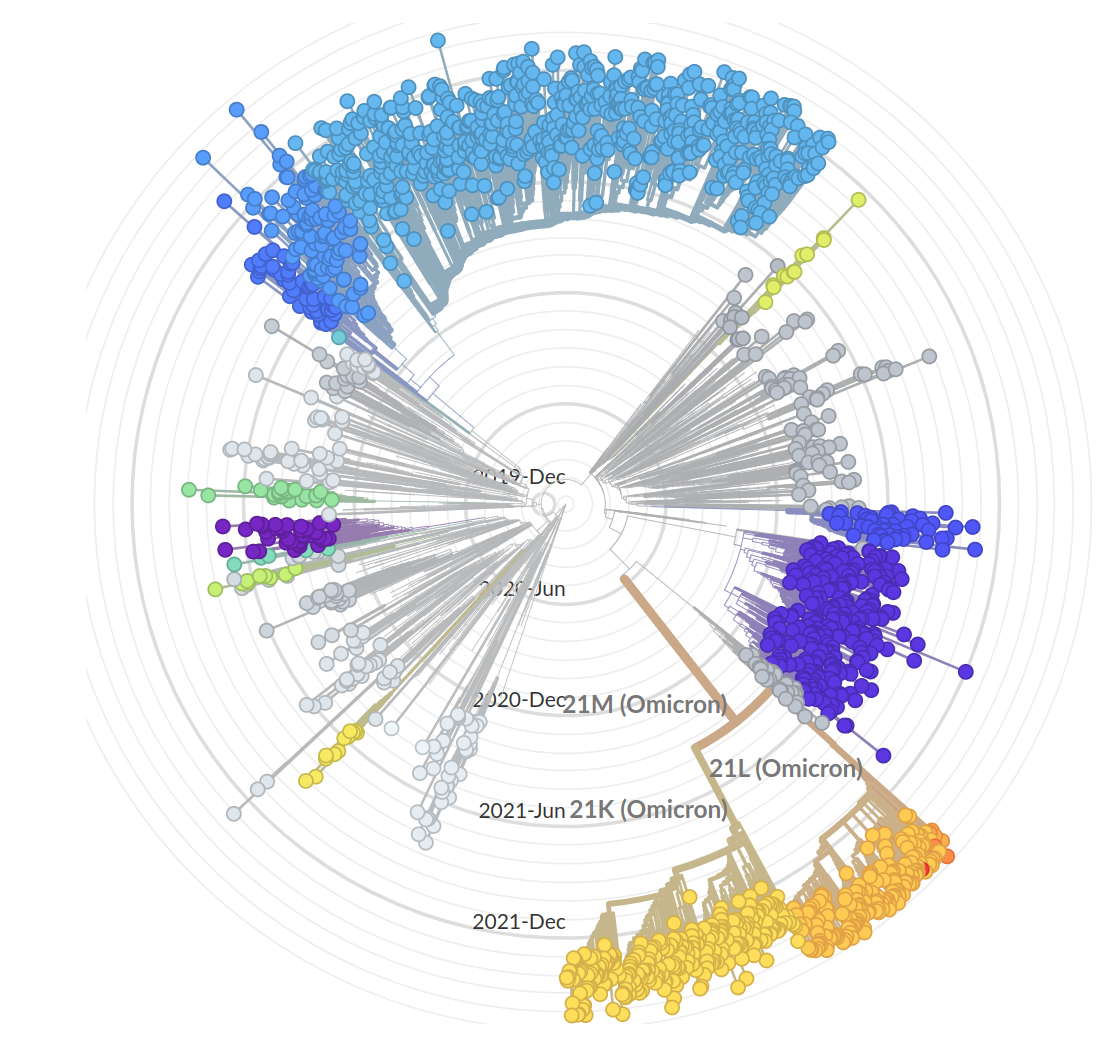
- Variant emergence likely through chronic infections
- Strong dichotomy (until 2022): dramatic changes between variants, slow and steady within variants
- Omicron variants are more dynamic
- With BA.4/5 and BA.2 subvariants, we start to see second generation variants
What is driving this?
Can we predict?
To predict, we need to quantify selection by immunity
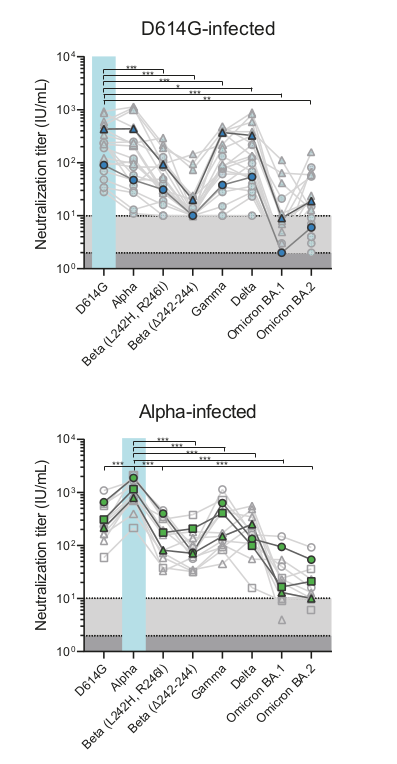
- Given a population immunity "landscape", how much escape is caused by which mutation?
- How variable are individual immune responses?
- How does exposure history shape neutralization of different variants?
- What is the contribution of chronic vs acute infections?
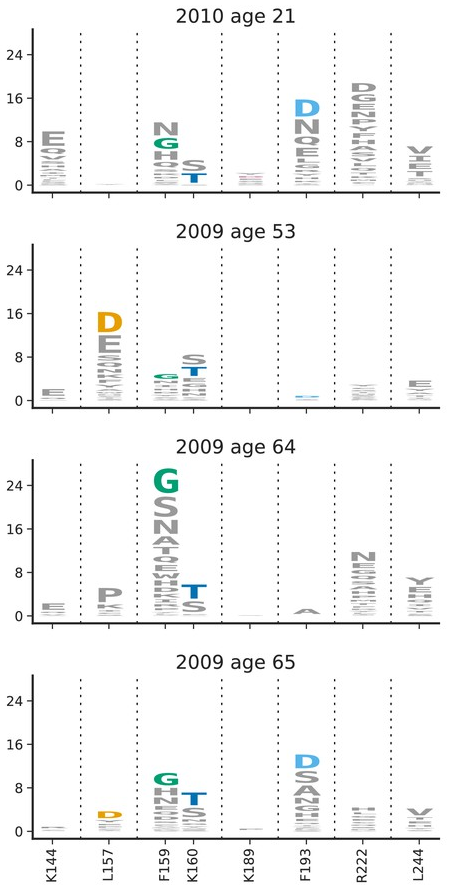
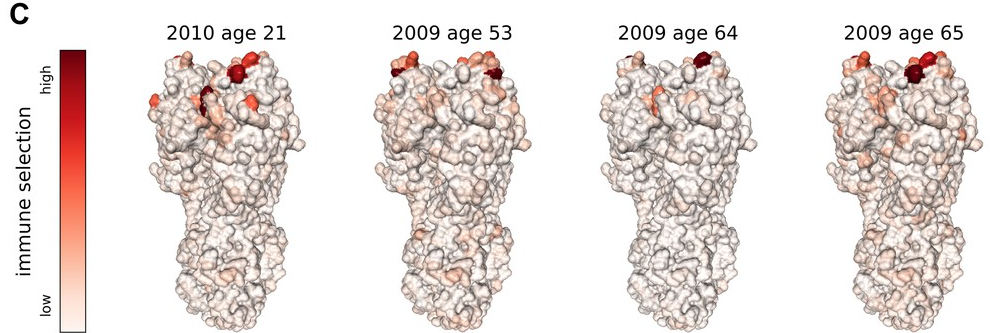
- Does escape during chronic infection mediate inter-individual escape? It did for Omicron, but will this stay that way?
- What is the contribution of chronic infections in other viruses?
Acknowledgments
- Fabio Zanini
- Jan Albert
- Johanna Brodin
- Christa Lanz
- Göran Bratt
- Lina Thebo
- Vadim Puller



Acknowledgments
- Emma Hodcroft
- Moira Zuber
- Inaki Comas
- Fernando Gonzales
- Tanja Stadler
- Sarah Nadeau
- Tim Vaughan
- Jesse Bloom
- David Veesler

Acknowledgments
Trevor Bedford and his lab -- terrific collaboration since 2014

especially James Hadfield, Emma Hodcroft, Ivan Aksamentov, Moira Zuber, John Huddleston, and Tom Sibley
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID

