Predictability of influenza virus evolution
Richard Neher
Biozentrum & SIB, University of Basel
slides at neherlab.org/202303_RoyalSociety.html
Human seasonal influenza viruses

Positive tests for influenza in the USA by week
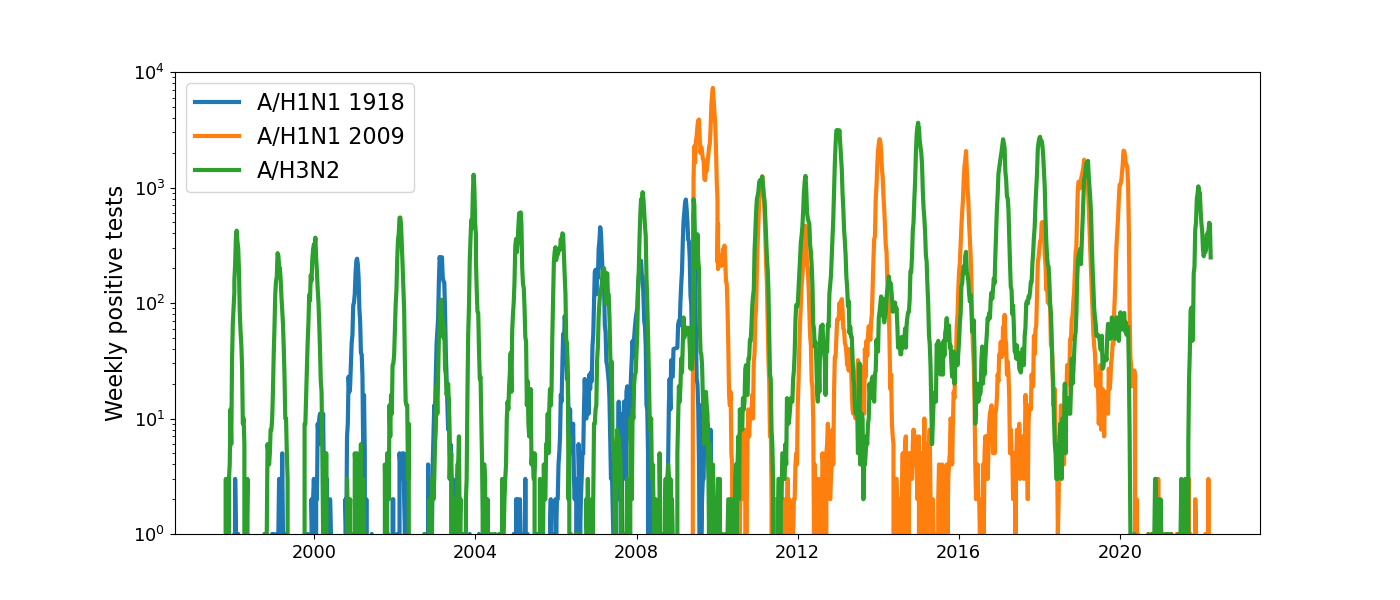 Data by the US CDC
Data by the US CDC
Genomic analysis to reconstruct pathogen spread and evolution
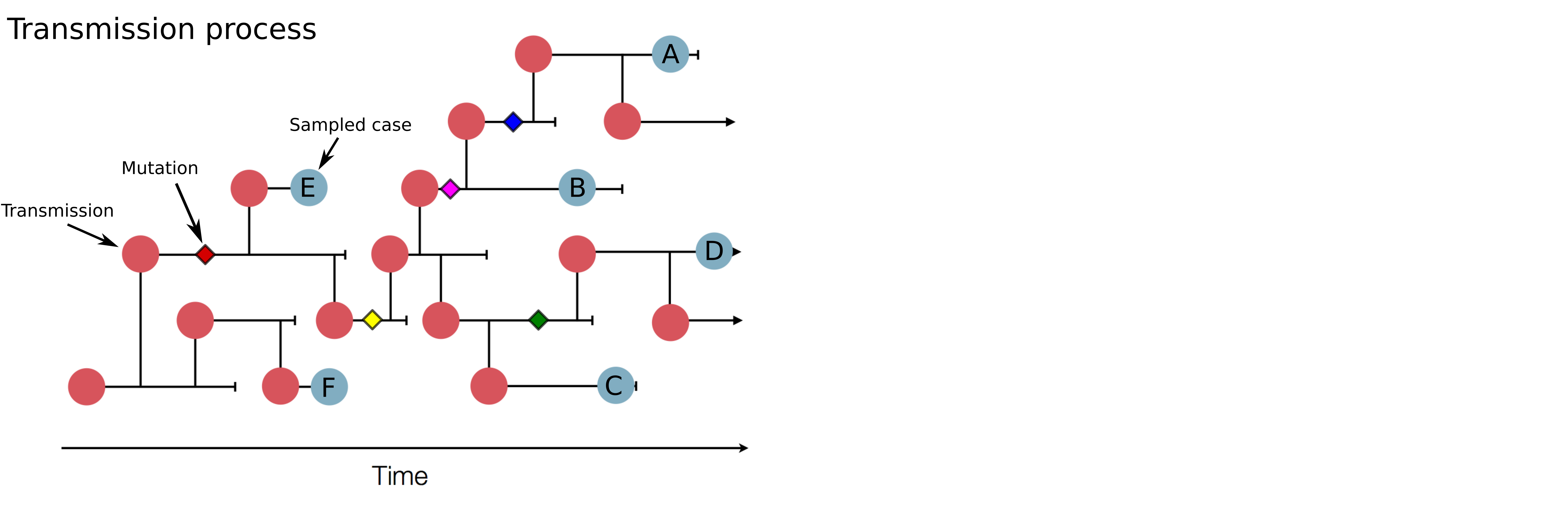
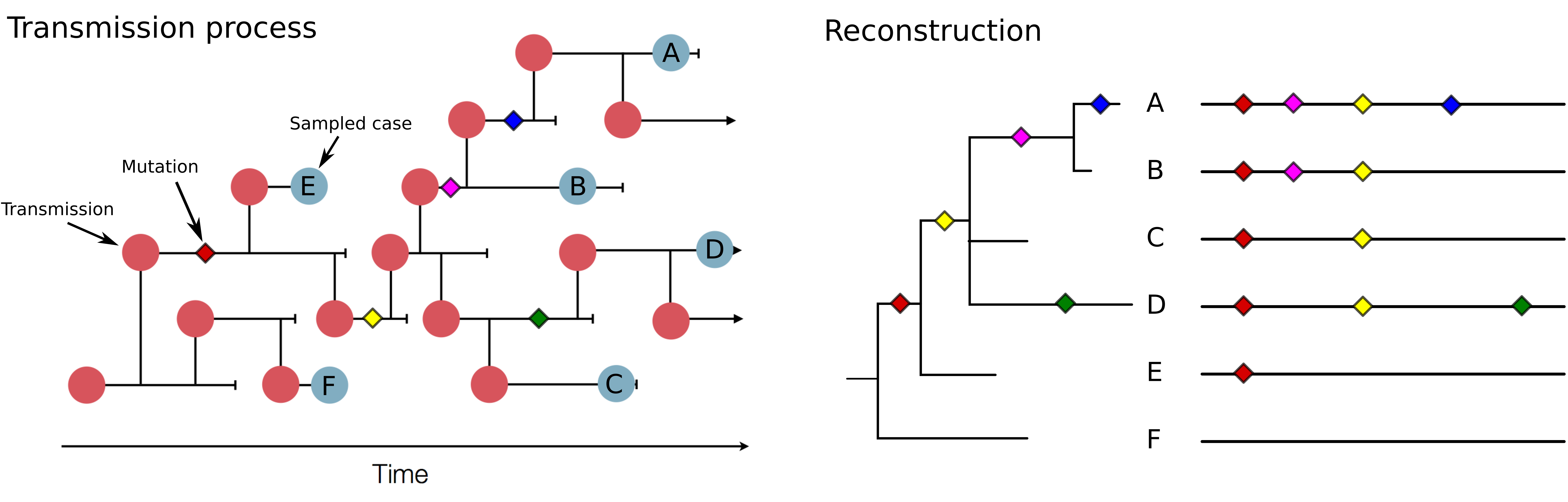
Link genotypic and phenotypic changes: immune escape, drug resistance, host adaptation
Influenza A/H3N2



- Influenza viruses evolve to avoid human immunity
- Vaccines need frequent updates

Vaccine strain selection schedule
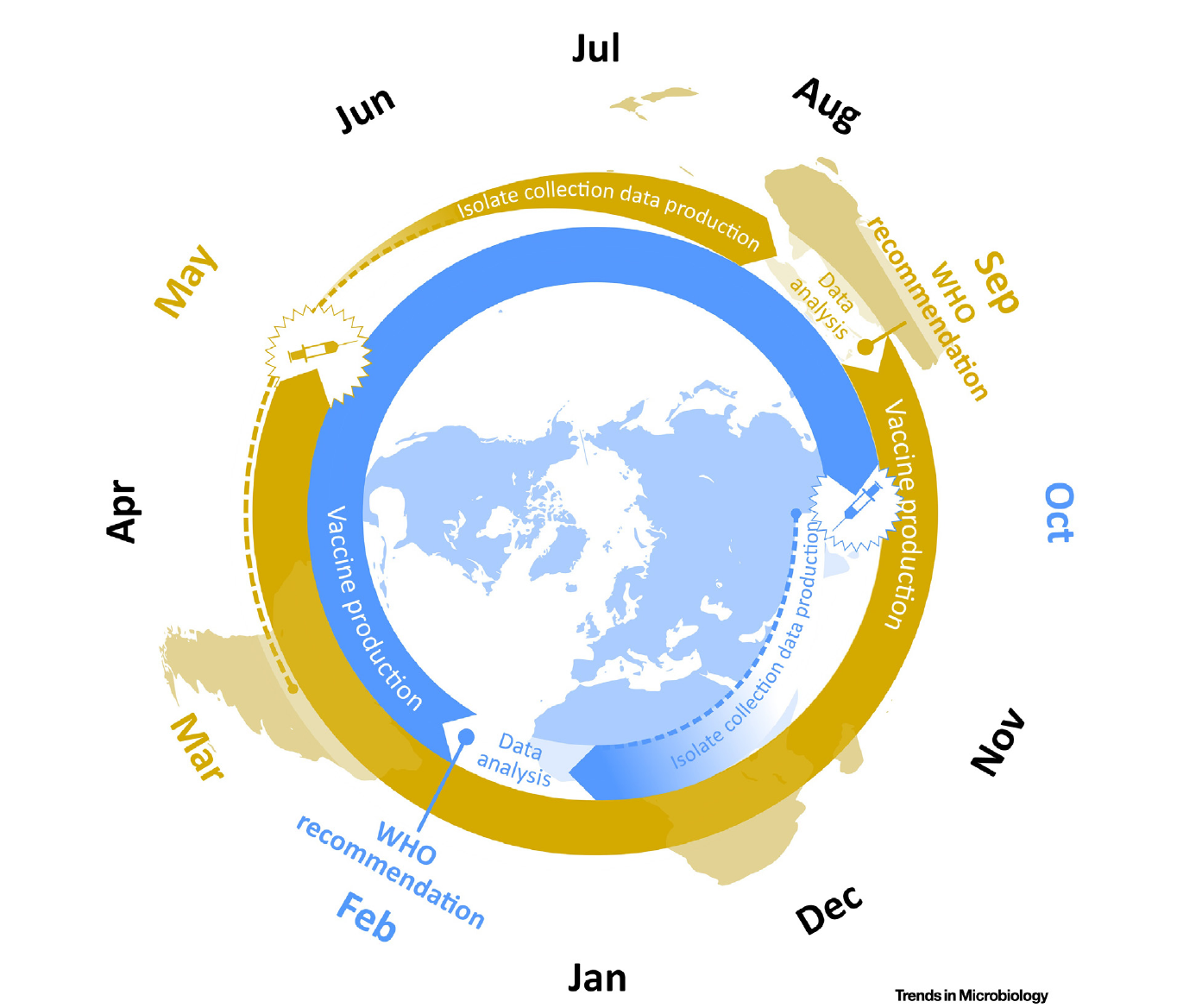 Klingen and McHardy, Trends in Microbiology
Klingen and McHardy, Trends in Microbiology
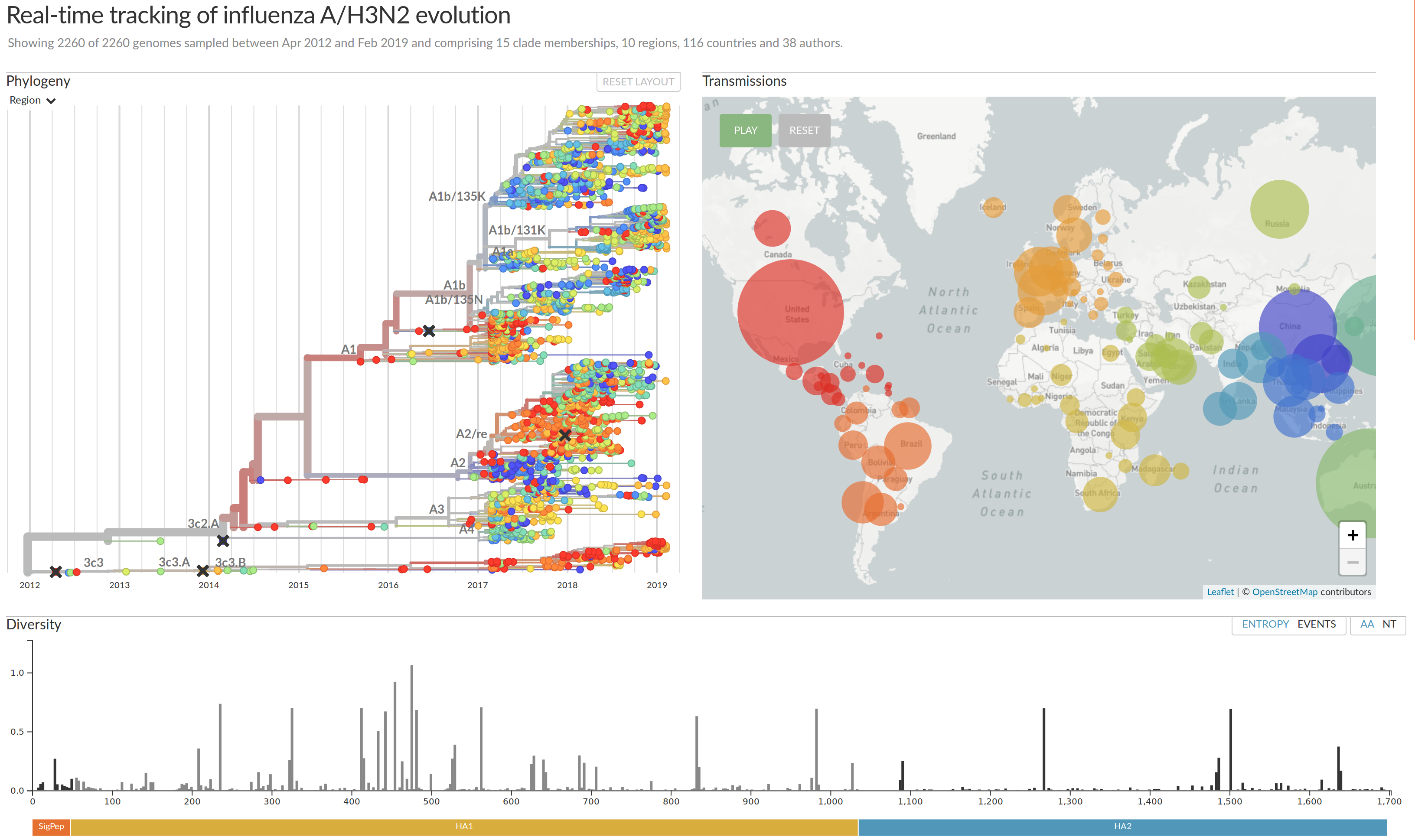
Beyond tracking: can we predict?
Can we pick a "winner"?
Traditional approach: evolutionary race of viruses
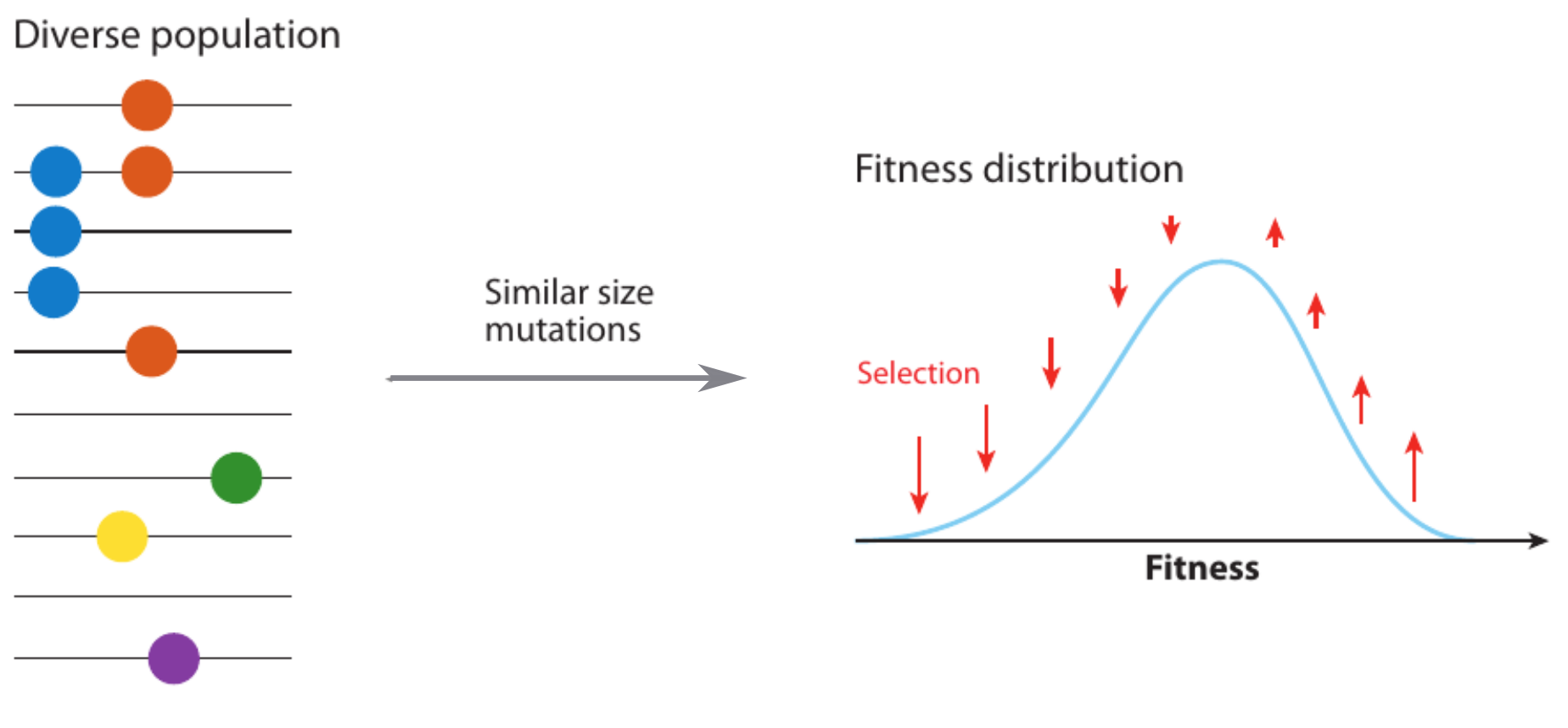
- Speed of adaptation is logarithmic in population size
- Single mutations are easy to find, many such mutation are needed for success
- Different models have universal emerging properties
Neutral/Kingman coalescent
strong selection
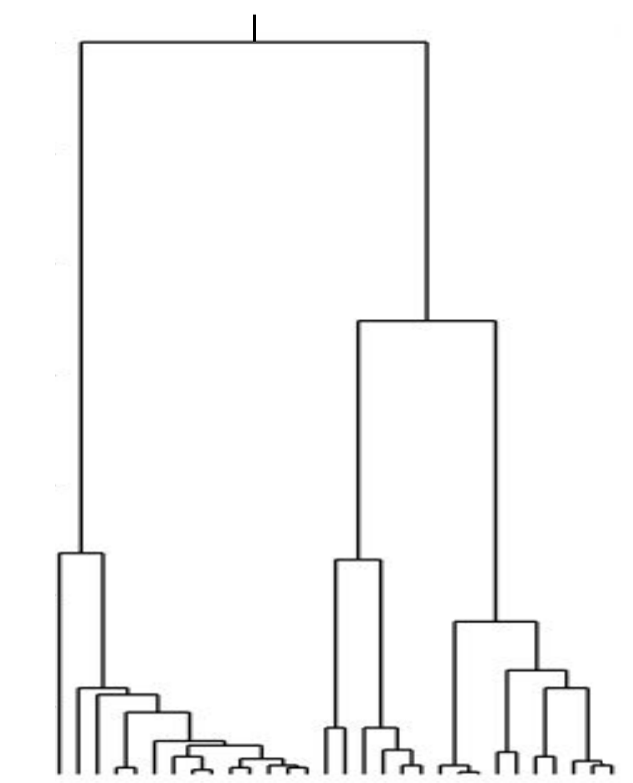

Bolthausen-Sznitman Coalescent
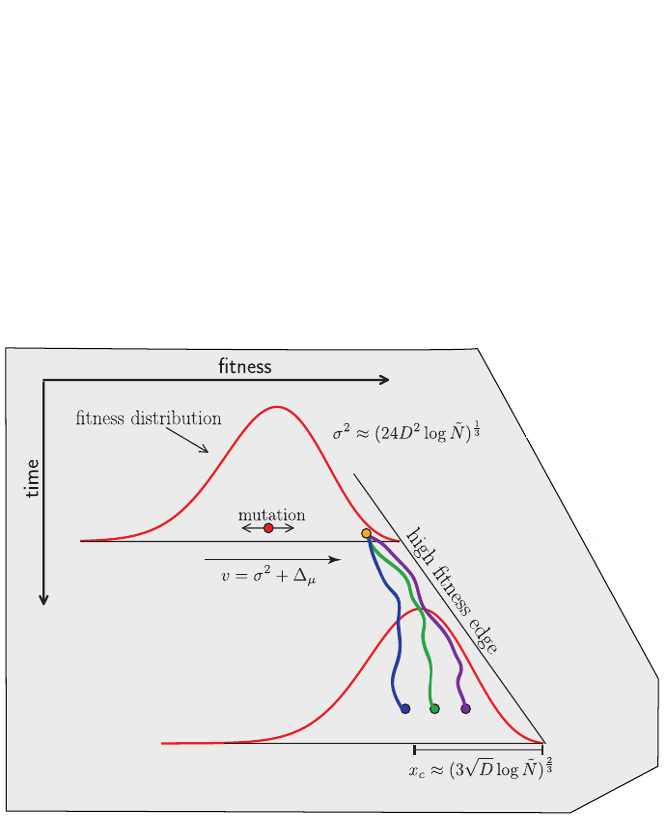
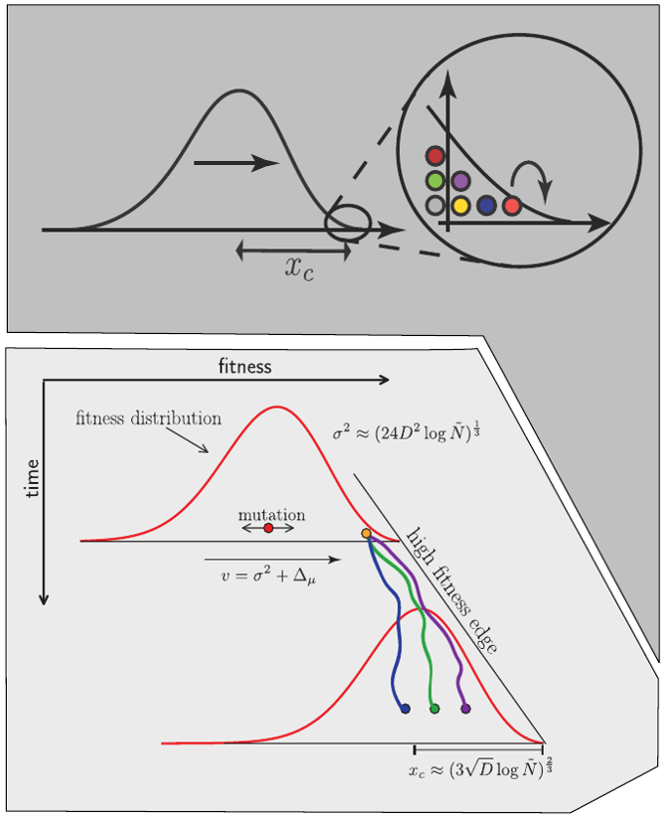
Burst in the tree ↔ high fitness
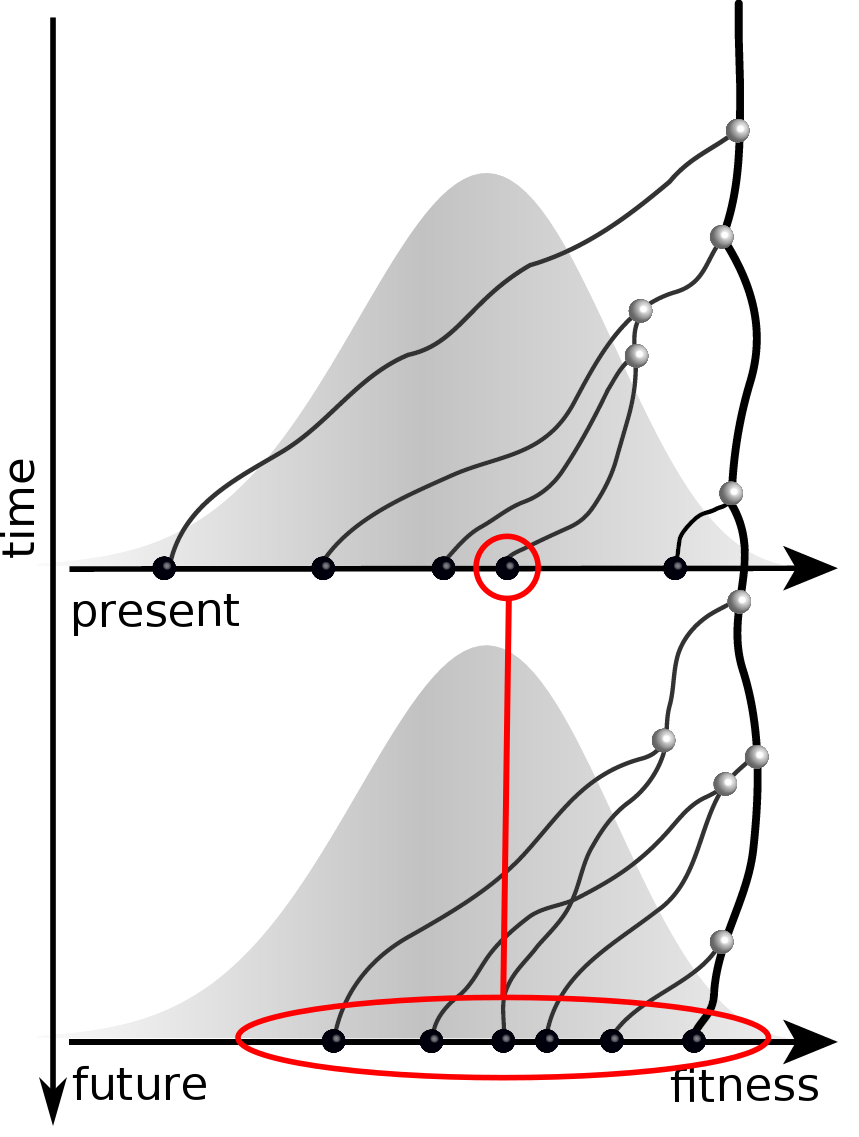
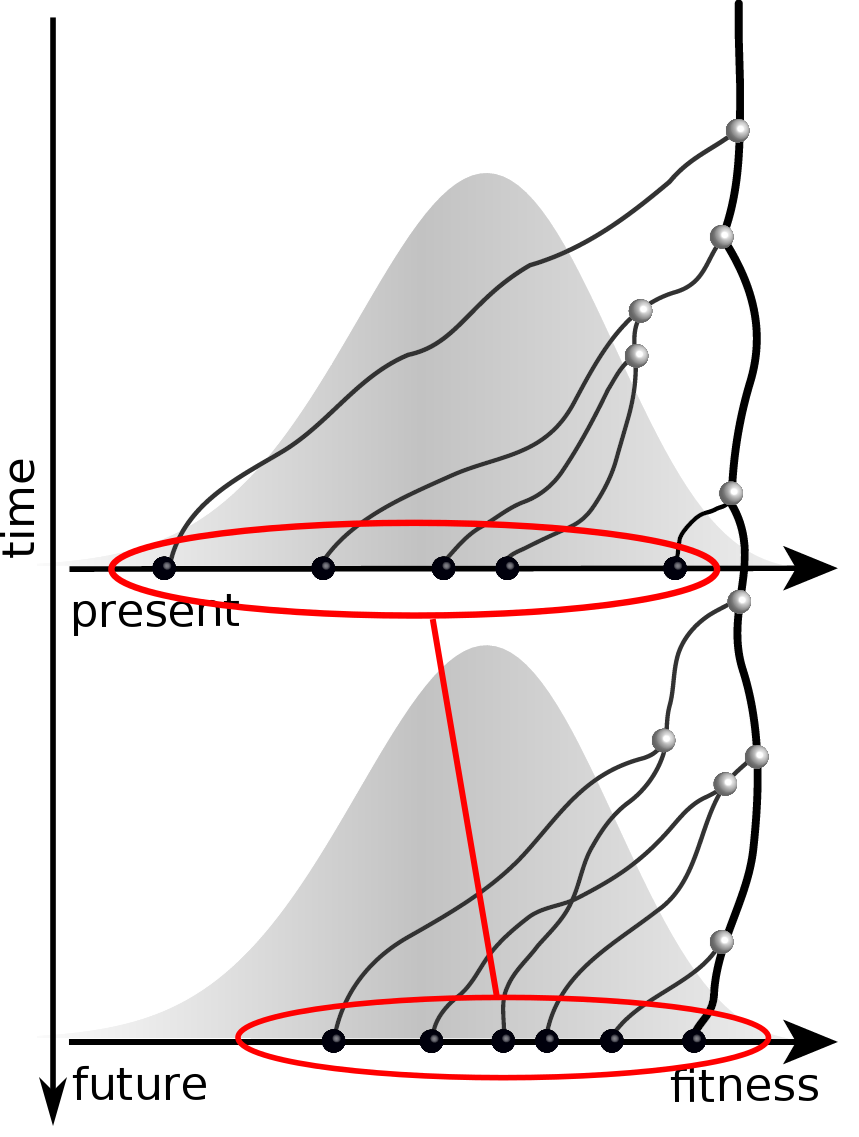
Predicting evolution

Given the branching pattern:
- can we predict fitness?
- pick the closest relative of the future?
Fitness inference from trees
$$P(\mathbf{x}|T) = \frac{1}{Z(T)} p_0(x_0) \prod_{i=0}^{n_{int}} g(x_{i_1}, t_{i_1}| x_i, t_i)g(x_{i_2}, t_{i_2}| x_i, t_i)$$
RN, Russell, Shraiman, eLife, 2014
Predicting an optimal representative
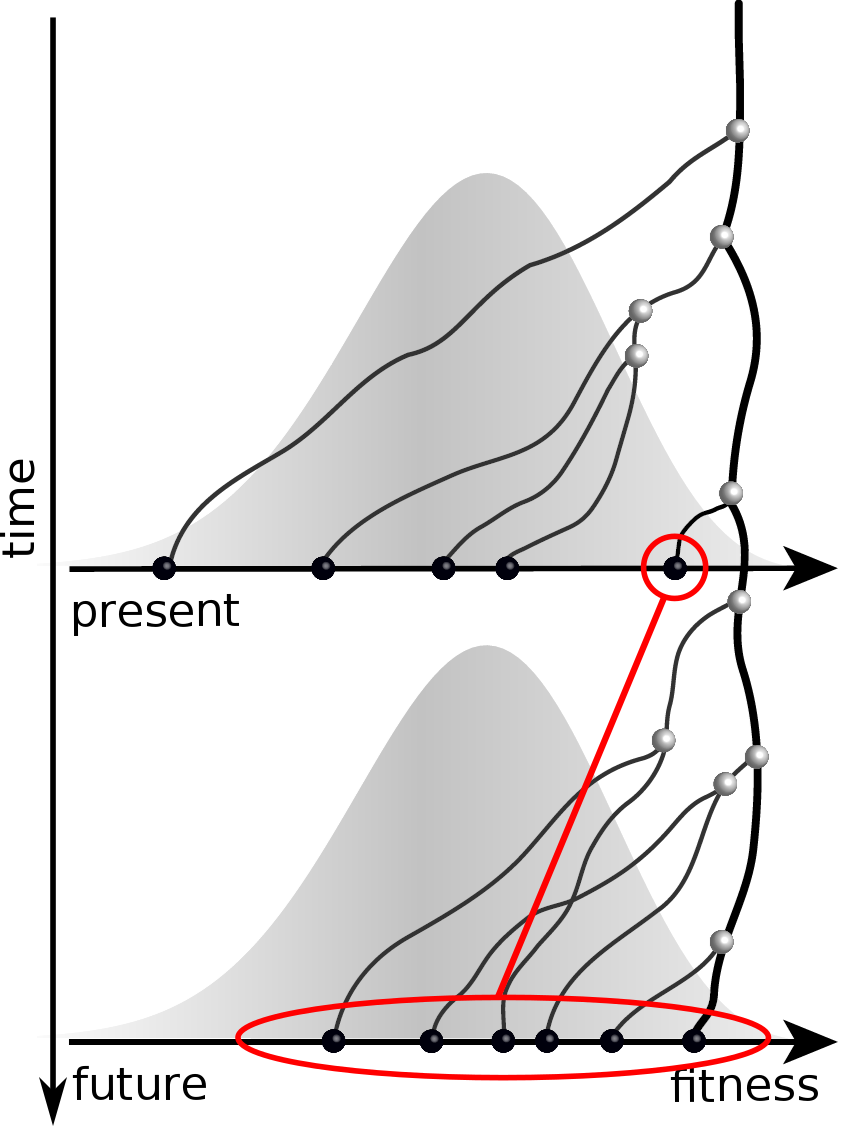
Moderate prediction success
- But is a random strain for previous years a sensible null?
- Does this work for the right reason?
Predicting the distribution in sequence space
Huddleston et al, eLife, 2020Follow up investigations show very modest predictability
- Optimal transport distances between prediction and observation -- less than one amino acid gained
Do A/H3N2 mutations have inertia?
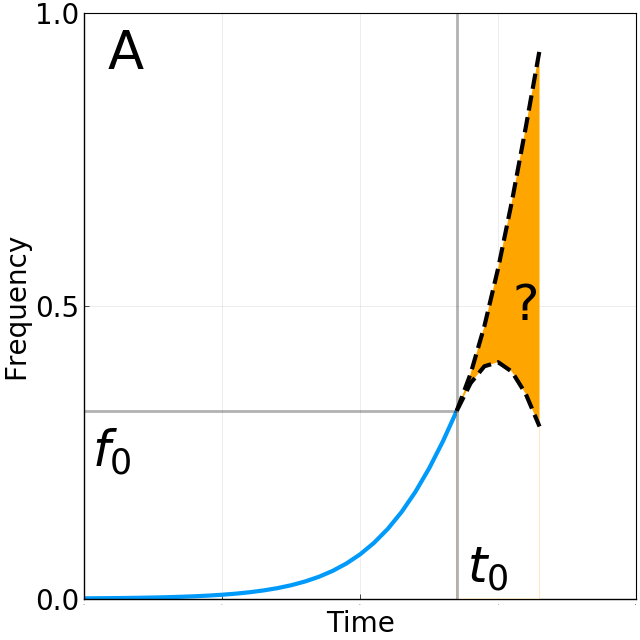
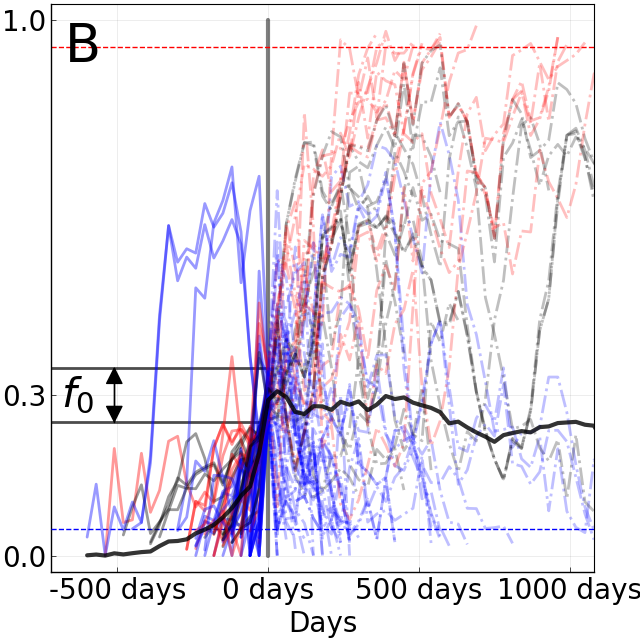
Complicated "Ecology" of host and pathogen
- Current approaches focus on the virus population
- Models are predicated on identifying a 'winner'
- Instead, dynamics might be slaved to host immunity:
→ exposure history and waning determine population immunity, viruses fill whatever niche there is
- For prediction, host data would be at least as important as viral dynamics
- Prediction horizon is limited to within season dynamics that equilibrates the host-pathogen immunity landscape
- Viral adaptations are niche specific and loose their benefit after equilibration.
Diversity in immune responses
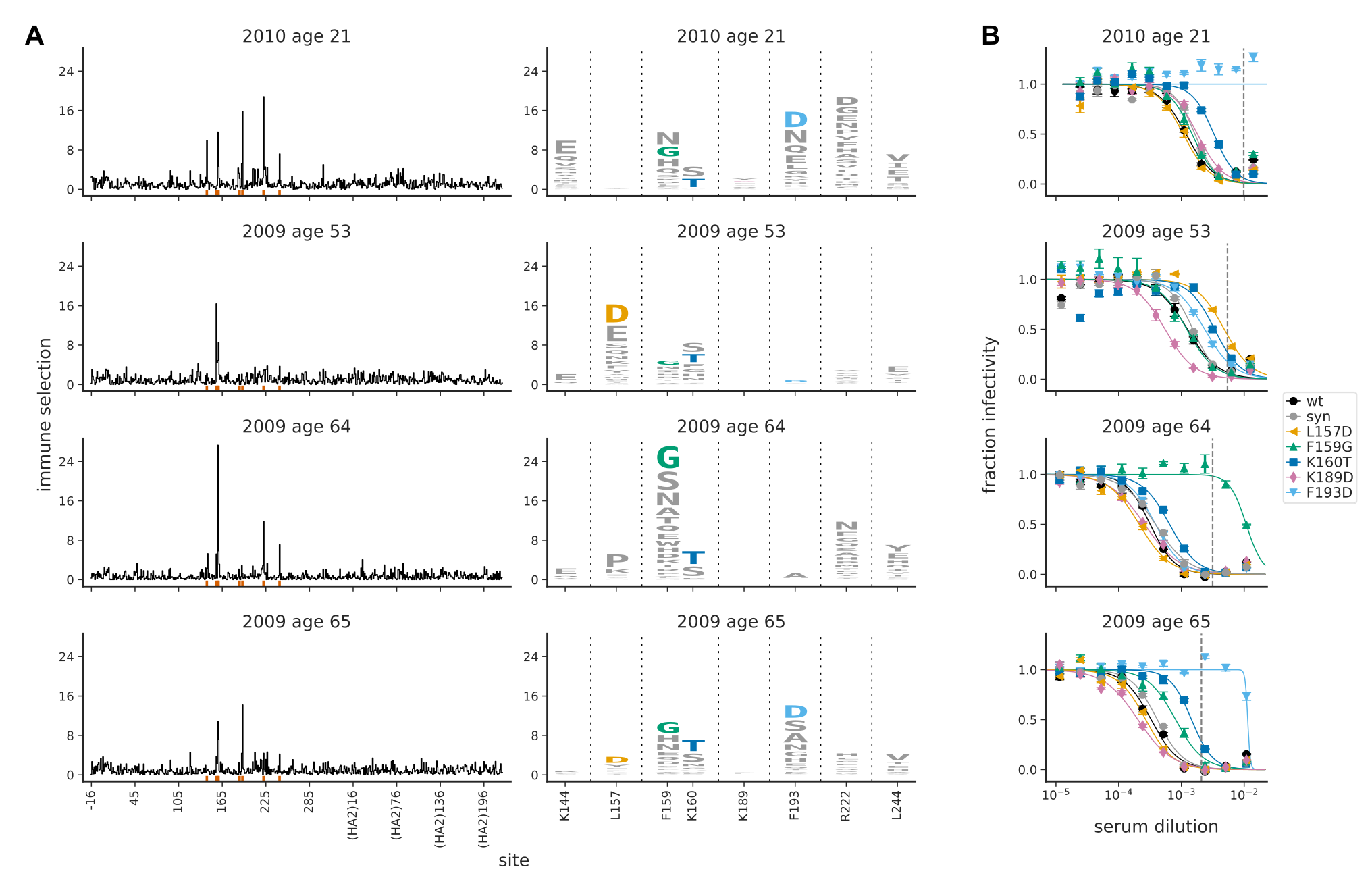
Influenza and Theory acknowledgments





- Boris Shraiman
- Colin Russell
- Trevor Bedford
- Pierre Barrat
- John Huddleston
- All the NICs and WHO CCs that provide influenza sequence data
- The WHO CCs in London and Atlanta for providing titer data

Acknowledgments
Trevor Bedford and his lab -- terrific collaboration since 2014

especially James Hadfield, Emma Hodcroft, Ivan Aksamentov, Cornelius Roemer, Moira Zuber, and John Huddleston
Data we analyze are contributed by scientists from all over the world
Data are shared and curated by GISAID


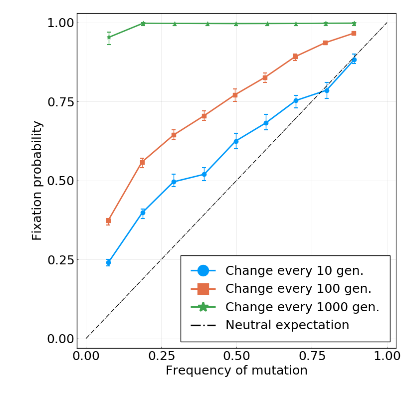
Fixation probability is quasi-neutral
A/H3N2 influenza
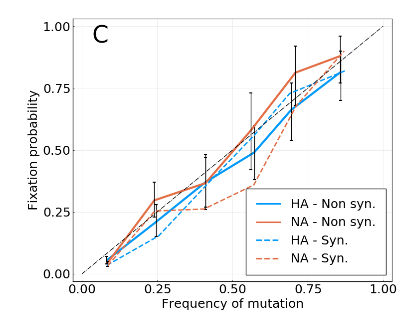
Simulations with increasing interference